|
chr1_-_26105477
|
1.551
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr2_-_37752272
|
1.503
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr6_-_72187168
|
1.429
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr14_-_22458220
|
1.421
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr18_-_21186107
|
1.417
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr14_-_22458199
|
1.382
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr14_-_22458134
|
1.345
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr19_-_45888259
|
1.235
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr17_-_7996409
|
1.233
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr8_-_9046460
|
1.226
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chrX_-_83644054
|
1.177
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr12_-_6103945
|
1.148
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr8_-_120033240
|
1.144
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr19_+_47593137
|
1.109
|
|
|
|
|
chr2_-_144991386
|
1.058
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr13_-_98428244
|
1.001
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr11_+_113672378
|
0.997
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr14_-_37795001
|
0.997
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr8_-_93184629
|
0.976
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_55533047
|
0.951
|
NM_022454
|
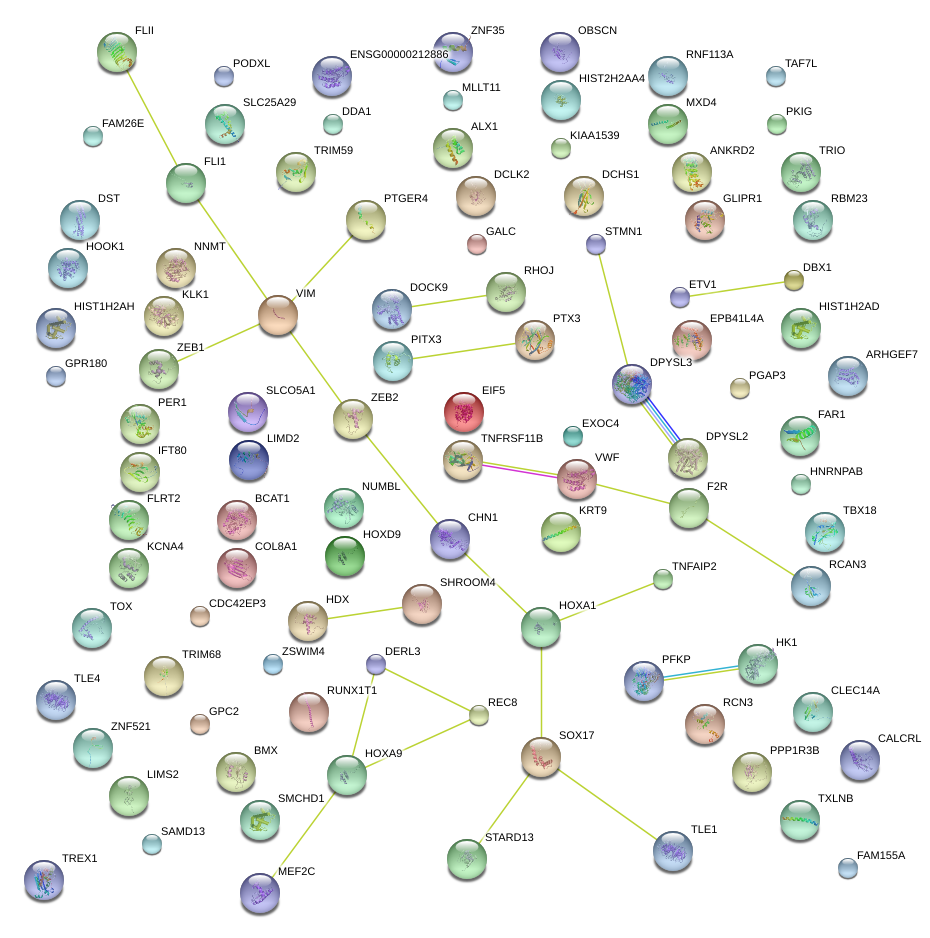
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr13_+_110653407
|
0.948
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr20_+_42644633
|
0.926
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr11_+_113672407
|
0.924
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr13_-_107317460
|
0.918
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr14_-_37795290
|
0.914
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr8_-_60194099
|
0.901
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chrX_+_15428820
|
0.899
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr14_-_87529600
|
0.899
|
NM_000153
NM_001037525
|
GALC
|
galactosylceramidase
|
|
chr9_+_81377419
|
0.897
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr13_-_32658215
|
0.890
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_37752731
|
0.886
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr11_-_4585947
|
0.883
|
NM_018073
|
TRIM68
|
tripartite motif containing 68
|
|
chr2_-_175578156
|
0.876
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr7_-_27171648
|
0.854
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_99612757
|
0.845
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr5_+_76047536
|
0.841
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr7_-_13995811
|
0.841
|
|
ETV1
|
ets variant 1
|
|
chr11_+_128068982
|
0.833
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr7_-_27171673
|
0.811
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr14_-_87529212
|
0.811
|
|
GALC
|
galactosylceramidase
|
|
chr12_+_84198015
|
0.809
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr19_+_13767207
|
0.805
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr2_-_128138435
|
0.800
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr10_+_70748581
|
0.789
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr10_+_17311242
|
0.788
|
|
VIM
|
vimentin
|
|
chr18_-_21185968
|
0.780
|
|
ZNF521
|
zinc finger protein 521
|
|
chr3_+_48482625
|
0.779
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr1_-_197173159
|
0.776
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr10_+_17311282
|
0.773
|
|
VIM
|
vimentin
|
|
chr7_+_148613304
|
0.772
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr14_-_87529222
|
0.771
|
|
GALC
|
galactosylceramidase
|
|
chr17_-_59817456
|
0.765
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr1_-_26105230
|
0.763
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr1_+_149299530
|
0.756
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_+_17312654
|
0.751
|
|
VIM
|
vimentin
|
|
chr1_+_149299504
|
0.749
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr14_+_23711044
|
0.746
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr11_+_13646727
|
0.743
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr22_-_22511173
|
0.734
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr17_-_59817575
|
0.724
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_+_3099708
|
0.721
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr1_+_84539876
|
0.721
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr13_+_94052022
|
0.718
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr11_+_13646806
|
0.717
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr10_+_22765996
|
0.717
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr3_+_44665220
|
0.703
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr17_-_59817654
|
0.702
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_+_31650069
|
0.699
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr5_-_88214720
|
0.695
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr10_+_17311283
|
0.691
|
|
VIM
|
vimentin
|
|
chr1_+_149299490
|
0.691
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr9_+_81377289
|
0.689
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr14_+_102659464
|
0.685
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr5_+_177564102
|
0.685
|
NM_004499
NM_031266
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr7_-_27171600
|
0.682
|
|
HOXA9
|
homeobox A9
|
|
chr11_+_128069165
|
0.682
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr2_-_188021117
|
0.678
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr10_+_17311307
|
0.677
|
|
VIM
|
vimentin
|
|
chr17_-_59817722
|
0.672
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr14_+_85066265
|
0.669
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr14_+_62740878
|
0.664
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr3_+_100840129
|
0.663
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr7_-_130891861
|
0.662
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_177564139
|
0.659
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr8_+_60194328
|
0.656
|
|
|
|
|
chr4_+_151218862
|
0.654
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr11_-_6633427
|
0.651
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr14_+_85066207
|
0.642
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr7_+_27102197
|
0.638
|
|
|
|
|
chr5_+_95093267
|
0.628
|
|
|
|
|
chr8_+_26491507
|
0.618
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_-_50403412
|
0.617
|
|
SHROOM4
|
shroom family member 4
|
|
chr17_-_59131210
|
0.608
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr5_+_14493905
|
0.607
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr7_-_27102049
|
0.601
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr6_-_56824371
|
0.601
|
|
DST
|
dystonin
|
|
chr14_+_23710924
|
0.600
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr6_+_116939500
|
0.598
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr5_-_111782637
|
0.597
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr9_-_35101592
|
0.596
|
|
KIAA1539
|
KIAA1539
|
|
chr19_+_54723044
|
0.595
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr19_+_17281216
|
0.586
|
NM_024050
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr6_-_85529884
|
0.586
|
|
TBX18
|
T-box 18
|
|
chr3_+_158637273
|
0.586
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr5_+_40715799
|
0.585
|
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr11_+_128069183
|
0.584
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr5_-_146813343
|
0.584
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr12_-_24946495
|
0.582
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr8_-_70907957
|
0.579
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr19_+_17281352
|
0.579
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr14_-_99842519
|
0.579
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr3_+_48482213
|
0.576
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr4_-_2233606
|
0.576
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_+_27222839
|
0.573
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr5_+_40715778
|
0.570
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr19_+_17281374
|
0.565
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr12_+_74160779
|
0.562
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr3_-_161650294
|
0.560
|
NM_173084
|
TRIM59
|
tripartite motif containing 59
|
|
chr8_-_70908130
|
0.558
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr6_-_139654807
|
0.558
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr14_+_102870301
|
0.557
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr3_+_25444753
|
0.555
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr1_+_45565126
|
0.554
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr7_+_150321771
|
0.554
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr19_-_51856129
|
0.550
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr9_-_35101534
|
0.548
|
|
KIAA1539
|
KIAA1539
|
|
chr11_+_128069203
|
0.545
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_-_161650219
|
0.539
|
|
TRIM59
|
tripartite motif containing 59
|
|
chr8_-_101187346
|
0.537
|
|
RGS22
|
regulator of G-protein signaling 22
|
|
chr19_-_6230797
|
0.535
|
|
|
|
|
chr1_-_68470585
|
0.534
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr7_+_27102508
|
0.532
|
|
|
|
|
chr7_-_5535726
|
0.532
|
|
ACTB
|
actin, beta
|
|
chr2_-_144991556
|
0.528
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr14_+_102870431
|
0.527
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr9_+_89302574
|
0.526
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr2_-_56266408
|
0.525
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr21_-_14840506
|
0.525
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr12_+_14409875
|
0.524
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr17_-_1478293
|
0.522
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr13_-_29937966
|
0.521
|
|
HMGB1
HMGB1P6
|
high-mobility group box 1
high-mobility group box 1 pseudogene 6
|
|
chr5_-_124108672
|
0.520
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr19_+_50086362
|
0.518
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr7_-_5535432
|
0.517
|
|
ACTB
|
actin, beta
|
|
chr5_+_95092384
|
0.516
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr11_+_13646839
|
0.514
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr4_-_2233504
|
0.513
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_-_56824672
|
0.512
|
|
DST
|
dystonin
|
|
chr12_-_21701985
|
0.511
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr15_-_46257611
|
0.509
|
|
MYEF2
|
myelin expression factor 2
|
|
chr5_-_136862317
|
0.509
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_+_13646822
|
0.506
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chrX_+_12903145
|
0.505
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr1_+_149299307
|
0.496
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr7_-_8268666
|
0.495
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr12_-_21702000
|
0.493
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr4_-_81212737
|
0.491
|
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr1_-_43692134
|
0.490
|
NM_001190880
NM_031207
|
HYI
|
hydroxypyruvate isomerase (putative)
|
|
chr7_+_26158257
|
0.490
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr13_-_100125028
|
0.488
|
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr12_-_21702021
|
0.486
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr17_-_44010543
|
0.485
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr10_+_17311347
|
0.484
|
|
VIM
|
vimentin
|
|
chr12_-_21702036
|
0.484
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr14_+_102870245
|
0.481
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr13_-_29938029
|
0.480
|
NM_002128
|
HMGB1
|
high-mobility group box 1
|
|
chr3_-_170347019
|
0.478
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr5_+_66160359
|
0.474
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr20_-_55718982
|
0.473
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr8_+_1759502
|
0.473
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr9_+_123501186
|
0.471
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr22_-_37481925
|
0.468
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr17_-_59818033
|
0.466
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr12_-_21702046
|
0.466
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr5_+_177564140
|
0.466
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr10_+_70748633
|
0.464
|
|
HK1
|
hexokinase 1
|
|
chr11_-_64247230
|
0.462
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr11_-_27679022
|
0.461
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_+_102870287
|
0.457
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr15_-_43457916
|
0.455
|
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr9_+_4480193
|
0.454
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr10_+_17311962
|
0.454
|
|
VIM
|
vimentin
|
|
chr7_+_134114957
|
0.450
|
|
CALD1
|
caldesmon 1
|
|
chr12_-_46499834
|
0.447
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr1_-_26105954
|
0.447
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr19_+_50086357
|
0.444
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr19_+_50086299
|
0.443
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr6_-_27222556
|
0.440
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr6_-_53321658
|
0.440
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr5_-_141318810
|
0.434
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr10_-_14686243
|
0.434
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr1_+_43539239
|
0.433
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr5_-_136862130
|
0.432
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr2_+_12775814
|
0.431
|
|
|
|
|
chr1_+_145479805
|
0.431
|
NM_004326
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr15_-_42274760
|
0.427
|
|
FRMD5
|
FERM domain containing 5
|
|
chr7_+_134114912
|
0.426
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_137099602
|
0.426
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr7_+_115637795
|
0.426
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr3_+_174598937
|
0.423
|
NM_014932
|
NLGN1
|
neuroligin 1
|