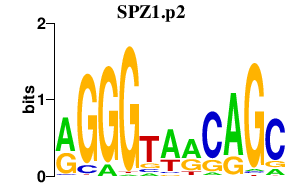
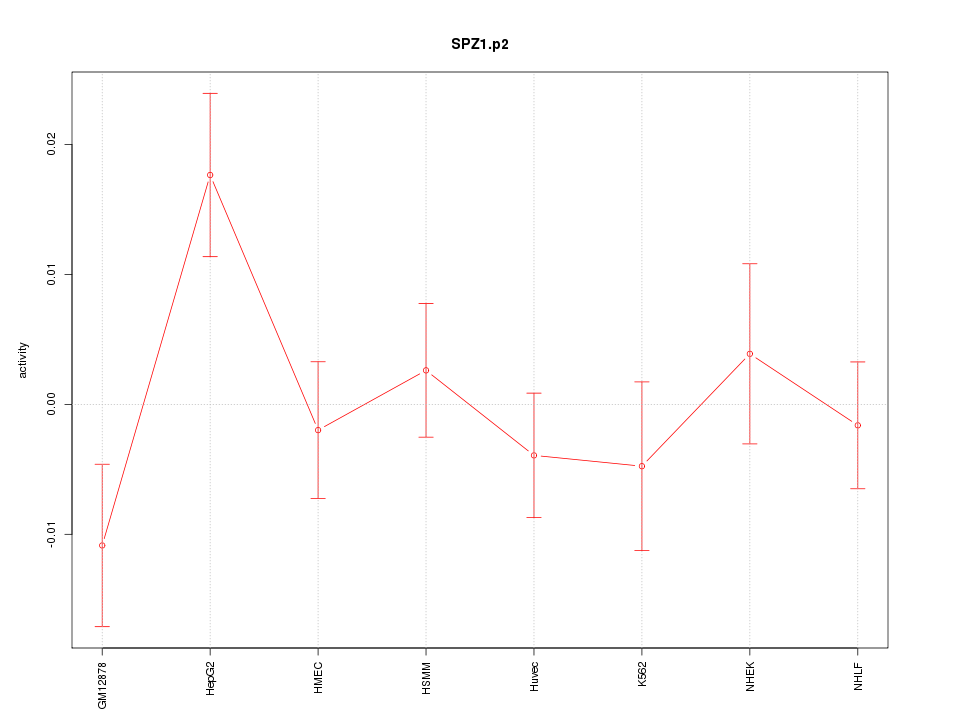
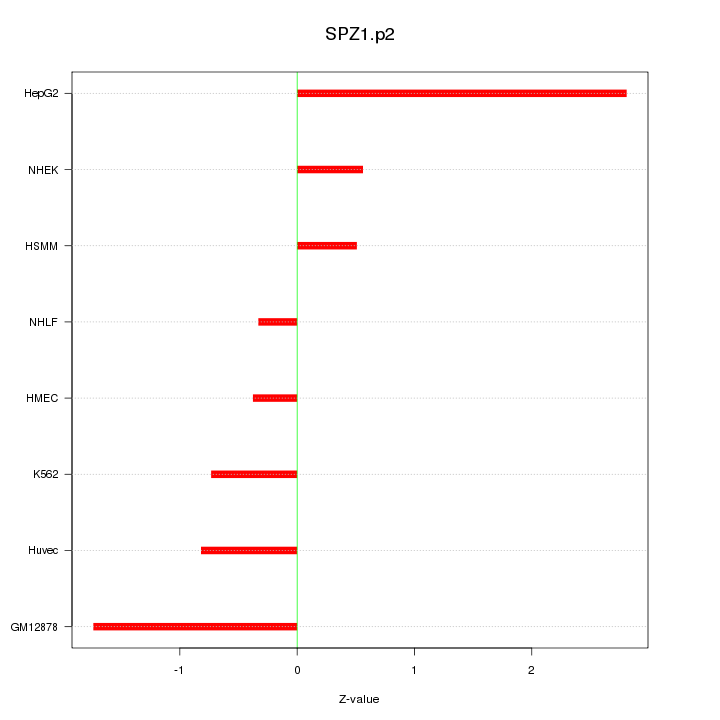
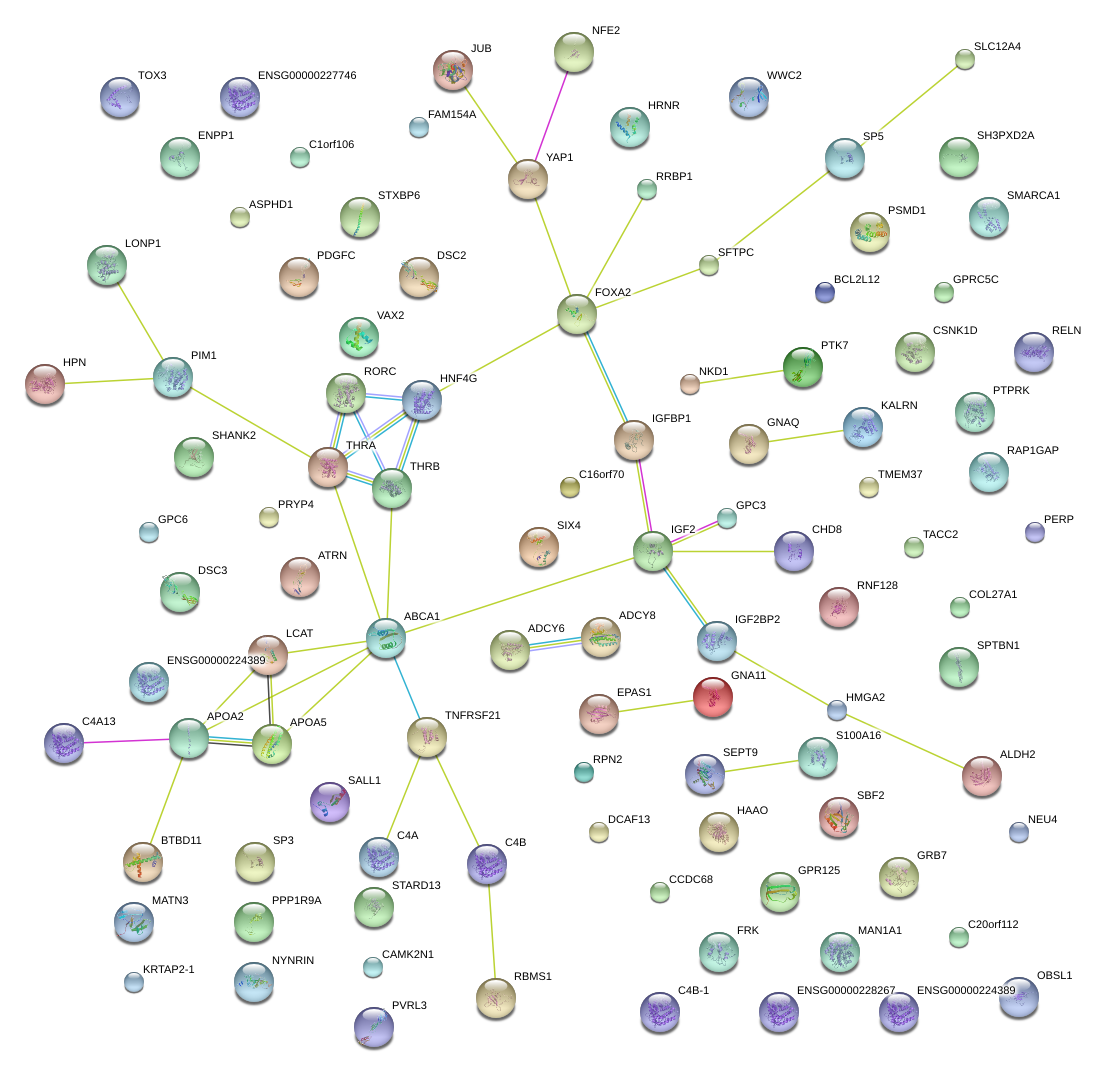
|
chr16_+_29819647
|
1.318
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr4_-_22126437
|
1.206
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr2_-_220144254
|
1.151
|
|
OBSL1
|
obscurin-like 1
|
|
chr4_+_184257337
|
1.140
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chrX_+_105856531
|
1.097
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr6_-_116488458
|
1.070
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr17_+_72795567
|
1.030
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chrX_-_128485121
|
1.029
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr1_-_20684858
|
0.963
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr16_-_66535457
|
0.934
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr7_+_94374862
|
0.926
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr20_-_22513050
|
0.905
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr7_+_45894480
|
0.874
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr3_-_187025438
|
0.853
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr2_+_54536780
|
0.836
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr18_-_26935883
|
0.835
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr10_+_123862430
|
0.808
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr12_+_64504835
|
0.784
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr3_-_187025501
|
0.780
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr6_-_128883125
|
0.773
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr14_-_22516271
|
0.771
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr9_-_106730157
|
0.740
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr4_-_187884721
|
0.705
|
|
|
|
|
chr20_-_42457717
|
0.696
|
|
|
|
|
chr1_-_159459999
|
0.692
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr9_+_115957537
|
0.688
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr1_-_150070849
|
0.677
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr20_-_30535045
|
0.670
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chrX_-_132947290
|
0.648
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chrX_-_132947141
|
0.647
|
|
GPC3
|
glypican 3
|
|
chr11_-_70641267
|
0.636
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_+_171280944
|
0.633
|
|
SP5
|
Sp5 transcription factor
|
|
chr11_-_2116775
|
0.625
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_-_66535923
|
0.623
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr6_-_47385205
|
0.615
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr16_+_29373365
|
0.612
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr17_-_36469869
|
0.606
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr12_-_52980914
|
0.598
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chrX_-_128485062
|
0.592
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr12_-_47468985
|
0.583
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr6_-_138231006
|
0.581
|
|
|
|
|
chr16_-_49742008
|
0.575
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr3_-_24511180
|
0.571
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr19_+_40223249
|
0.569
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr1_+_199130571
|
0.566
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr20_-_17610704
|
0.558
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_+_125786195
|
0.554
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr7_-_103417198
|
0.550
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr20_-_22512893
|
0.543
|
|
FOXA2
|
forkhead box A2
|
|
chr2_+_119905812
|
0.540
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr12_+_106236281
|
0.539
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr16_-_51138306
|
0.532
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr14_-_24588916
|
0.525
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr6_+_132170888
|
0.523
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_-_47385313
|
0.520
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr3_-_24511456
|
0.518
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr14_+_23937766
|
0.515
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr13_-_33148899
|
0.509
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_77824849
|
0.507
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr10_-_105411339
|
0.505
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr9_+_115958051
|
0.497
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr6_+_32090516
|
0.495
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr18_-_50777634
|
0.492
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr2_+_70981223
|
0.489
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr6_-_138470160
|
0.488
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr13_+_92677012
|
0.485
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr16_+_49139694
|
0.482
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr2_-_20075889
|
0.480
|
|
MATN3
|
matrilin 3
|
|
chr6_+_43151922
|
0.479
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr11_+_2116486
|
0.477
|
|
|
|
|
chr20_+_3399615
|
0.477
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr14_-_60260780
|
0.475
|
|
SIX4
|
SIX homeobox 4
|
|
chr2_-_20075929
|
0.474
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr3_+_112273352
|
0.461
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr2_+_70981233
|
0.457
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr20_-_17610828
|
0.455
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr16_+_65701428
|
0.453
|
|
|
|
|
chr20_-_22514092
|
0.452
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr11_-_116168316
|
0.450
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr2_-_174538197
|
0.449
|
|
SP3
|
Sp3 transcription factor
|
|
chr4_-_158111995
|
0.446
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr9_-_79835970
|
0.444
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr16_+_65701284
|
0.442
|
NM_025187
|
C16orf70
|
chromosome 16 open reading frame 70
|
|
chr17_+_69939806
|
0.441
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr11_+_101488380
|
0.440
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr11_-_10272256
|
0.440
|
NM_030962
|
SBF2
|
SET binding factor 2
|
|
chr1_-_151848039
|
0.439
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr1_-_21868380
|
0.438
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr4_-_158111504
|
0.436
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_+_242400702
|
0.434
|
NM_001167599
NM_001167602
NM_080741
|
NEU4
|
sialidase 4
|
|
chr2_+_46378405
|
0.430
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr14_-_20975241
|
0.429
|
NM_020920
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr17_+_35147697
|
0.428
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr2_-_42873179
|
0.425
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr6_-_119712596
|
0.424
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr2_-_161058120
|
0.421
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr6_+_37245860
|
0.420
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr9_-_106730256
|
0.420
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr20_+_35240844
|
0.418
|
NM_001135771
NM_002951
|
RPN2
|
ribophorin II
|
|
chr8_+_76482661
|
0.417
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr6_-_47385470
|
0.416
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr22_-_19122047
|
0.415
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr15_-_72804763
|
0.413
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr3_+_37878647
|
0.413
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr17_-_33178987
|
0.413
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr3_-_48445719
|
0.412
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr1_-_24307514
|
0.411
|
|
MYOM3
|
myomesin family, member 3
|
|
chr10_-_101680645
|
0.411
|
|
DNMBP
|
dynamin binding protein
|
|
chr19_-_6671585
|
0.410
|
NM_000064
|
C3
|
complement component 3
|
|
chr13_-_74953873
|
0.408
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr7_+_29200850
|
0.408
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr2_-_101134146
|
0.407
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr12_+_55808696
|
0.407
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr1_-_11909042
|
0.406
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr20_+_35241109
|
0.404
|
|
RPN2
|
ribophorin II
|
|
chr9_-_85342868
|
0.401
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr18_+_30812205
|
0.398
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr9_+_70926009
|
0.398
|
NM_001170414
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr12_+_6290359
|
0.397
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr20_+_35241161
|
0.397
|
|
RPN2
|
ribophorin II
|
|
chr6_-_24466258
|
0.395
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr17_+_33761234
|
0.394
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr16_+_66438348
|
0.391
|
|
NUTF2
|
nuclear transport factor 2
|
|
chr3_+_30622905
|
0.388
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr10_-_105604942
|
0.388
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr3_-_189354510
|
0.387
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr17_-_33179118
|
0.387
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr16_-_49742651
|
0.386
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr11_-_2116501
|
0.384
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_+_14145051
|
0.383
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr7_+_29200636
|
0.382
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr16_+_28741861
|
0.380
|
|
ATXN2L
|
ataxin 2-like
|
|
chr14_-_22974707
|
0.378
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr20_+_35240936
|
0.377
|
|
RPN2
|
ribophorin II
|
|
chr14_-_23686269
|
0.377
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr11_-_2116360
|
0.375
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_-_40858775
|
0.374
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr5_+_80292313
|
0.372
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr12_+_6290127
|
0.372
|
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr7_-_37923040
|
0.372
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr14_-_20636337
|
0.370
|
|
ZNF219
|
zinc finger protein 219
|
|
chr14_-_37133927
|
0.370
|
|
FOXA1
|
forkhead box A1
|
|
chr5_+_80292213
|
0.370
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chrX_-_128485134
|
0.365
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr2_-_73193581
|
0.365
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr2_-_161058550
|
0.362
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr2_-_220144427
|
0.361
|
|
OBSL1
|
obscurin-like 1
|
|
chr10_-_71753684
|
0.359
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr16_+_28785087
|
0.357
|
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr16_-_52877868
|
0.354
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr12_+_6363482
|
0.354
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr2_-_36384
|
0.353
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr6_+_108988797
|
0.351
|
|
FOXO3
|
forkhead box O3
|
|
chr10_+_114700775
|
0.351
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr14_+_34521945
|
0.350
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr2_-_220144511
|
0.349
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr14_+_34521854
|
0.347
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr16_+_66438285
|
0.346
|
NM_005796
|
NUTF2
|
nuclear transport factor 2
|
|
chr2_+_219780192
|
0.344
|
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr17_-_67628523
|
0.342
|
|
FLJ37644
|
hypothetical LOC400618
|
|
chr10_-_118419459
|
0.342
|
NM_144661
|
C10orf82
|
chromosome 10 open reading frame 82
|
|
chr11_+_45900731
|
0.341
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr19_+_6415259
|
0.341
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr17_-_38978253
|
0.339
|
|
ETV4
|
ets variant 4
|
|
chr4_-_158111916
|
0.338
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr5_+_129268352
|
0.337
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr3_+_9933763
|
0.337
|
NM_032732
NM_153460
NM_153461
|
IL17RC
|
interleukin 17 receptor C
|
|
chr2_-_73193566
|
0.333
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr17_-_53387493
|
0.332
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr19_+_50041387
|
0.331
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr21_-_36774257
|
0.331
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr5_-_172689111
|
0.329
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr11_-_47164514
|
0.328
|
NM_001184974
NM_016223
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr19_+_49972965
|
0.327
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr18_+_3439972
|
0.324
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr19_+_50041400
|
0.324
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr20_-_25514137
|
0.324
|
NM_025176
|
NINL
|
ninein-like
|
|
chr2_-_160972606
|
0.323
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr2_-_11402150
|
0.323
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr2_+_113119985
|
0.322
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr22_-_22970850
|
0.322
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr15_-_81412354
|
0.321
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr1_-_56817802
|
0.320
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_+_970613
|
0.320
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr3_-_147361465
|
0.319
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr20_-_47238082
|
0.317
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr11_+_69165159
|
0.317
|
|
CCND1
|
cyclin D1
|
|
chr12_+_51730101
|
0.317
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_-_159459814
|
0.315
|
|
APOA2
|
apolipoprotein A-II
|
|
chr7_+_989321
|
0.314
|
NM_017781
|
CYP2W1
|
cytochrome P450, family 2, subfamily W, polypeptide 1
|
|
chr12_-_52980984
|
0.312
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr17_-_53420595
|
0.312
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr17_+_44429748
|
0.311
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr20_+_3399727
|
0.308
|
|
ATRN
|
attractin
|
|
chr19_+_50041424
|
0.306
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr17_-_38978772
|
0.305
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr17_-_77602493
|
0.304
|
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr2_+_219779737
|
0.302
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr4_-_118226119
|
0.300
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr12_+_48784057
|
0.299
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|