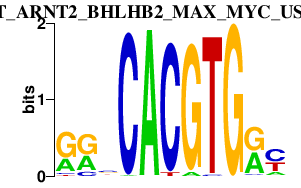
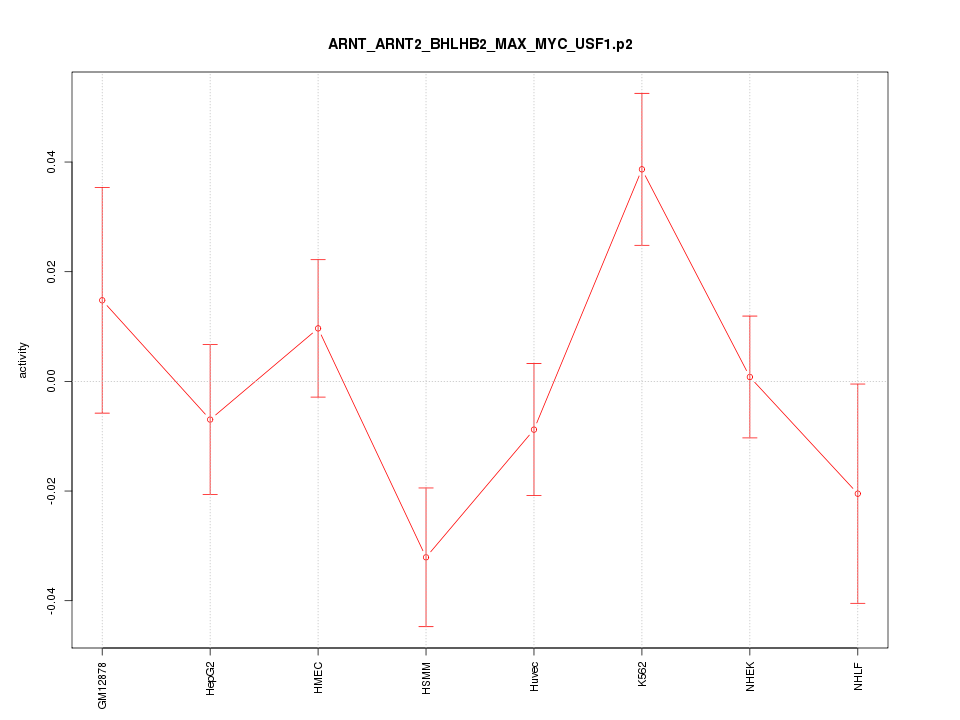
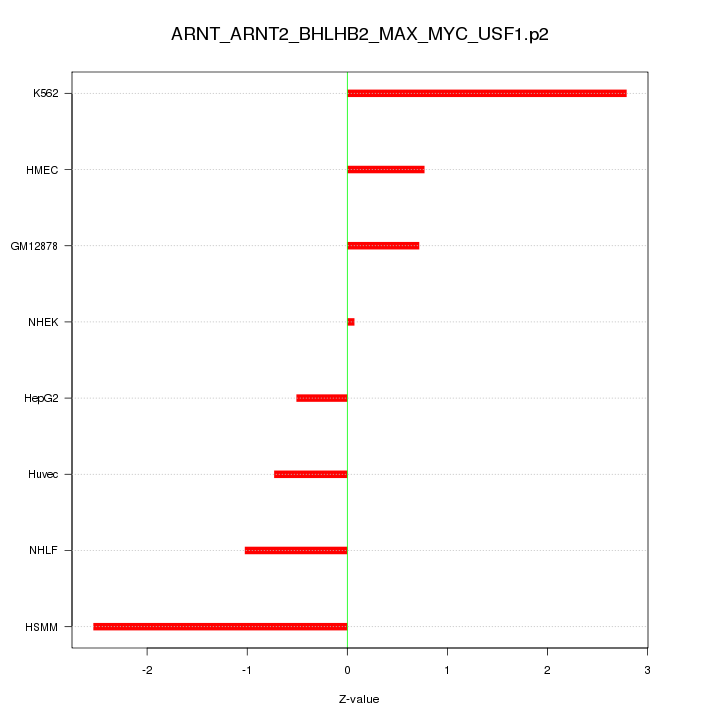
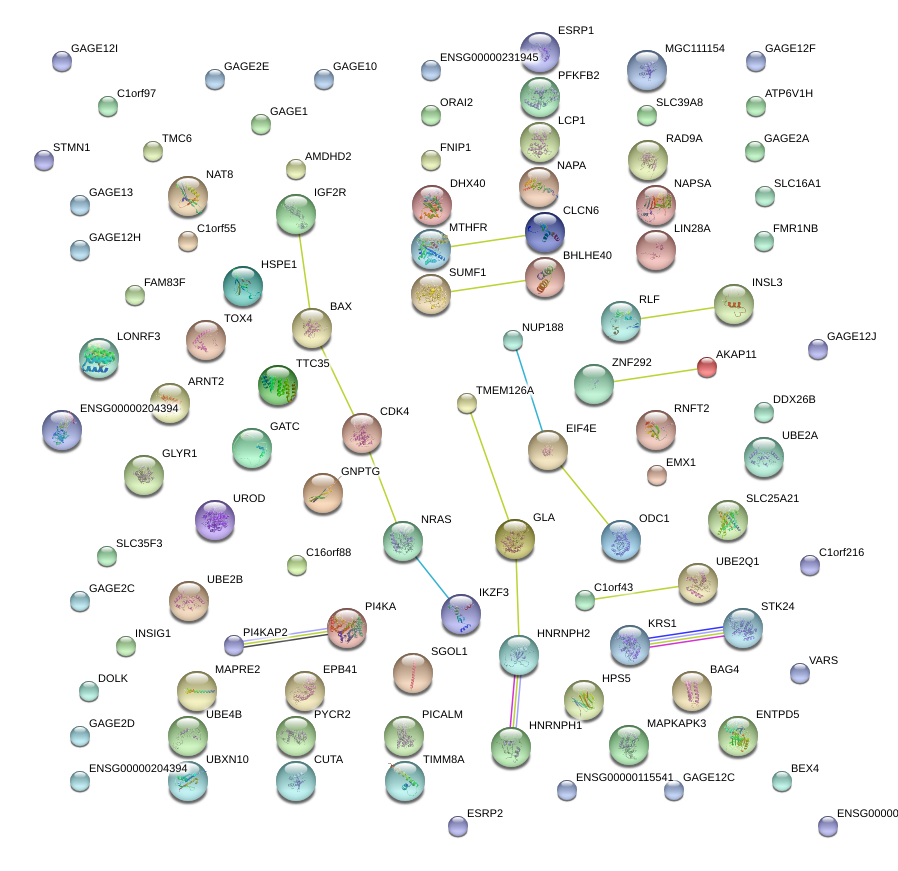
|
chr17_-_73636362
|
2.335
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr12_-_56432385
|
1.963
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr8_+_142471290
|
1.713
|
|
|
|
|
chr1_+_11788827
|
1.696
|
|
CLCN6
|
chloride channel 6
|
|
chr17_-_53764867
|
1.667
|
|
|
|
|
chrX_+_100549869
|
1.467
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_100549861
|
1.395
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr17_-_73636386
|
1.361
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr1_-_152797707
|
1.329
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chrX_+_100549839
|
1.293
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr16_+_2510323
|
1.286
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chrX_+_100549885
|
1.281
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_-_100490329
|
1.270
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr11_-_85457469
|
1.269
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr3_-_4483952
|
1.265
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr1_+_20385164
|
1.228
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr1_-_26105954
|
1.227
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr11_-_85457753
|
1.215
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chrX_+_102356659
|
1.213
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr1_-_26105477
|
1.196
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr11_-_85457736
|
1.156
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr6_-_33493792
|
1.144
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_-_11788666
|
1.141
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chrX_-_100549561
|
1.138
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr16_+_1341937
|
1.095
|
|
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit
|
|
chr3_-_4483928
|
1.054
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_-_4483955
|
1.036
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr4_-_100069265
|
1.017
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chrX_+_117992603
|
1.016
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr1_-_224178600
|
1.015
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_-_224253678
|
0.993
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr1_+_11788793
|
0.989
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chrX_+_134482212
|
0.968
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr12_-_56432292
|
0.968
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_+_111544469
|
0.959
|
|
|
|
|
chr1_-_113300351
|
0.945
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr9_-_130749604
|
0.917
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr16_-_4837266
|
0.914
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr1_+_45250475
|
0.912
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr12_-_56432377
|
0.909
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_+_10015736
|
0.907
|
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr1_-_224253650
|
0.884
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr6_+_87921980
|
0.876
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chrX_+_130985316
|
0.875
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr6_-_31871394
|
0.870
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr1_-_224253610
|
0.868
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr1_-_113300138
|
0.861
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr1_+_29113589
|
0.859
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_-_224253642
|
0.858
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr12_+_115660468
|
0.855
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr13_+_41744255
|
0.854
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr8_-_54918099
|
0.851
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr9_+_130749791
|
0.841
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr1_+_40399642
|
0.827
|
|
RLF
|
rearranged L-myc fusion
|
|
chr4_-_100069303
|
0.818
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chrX_+_117992623
|
0.815
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr6_+_160310118
|
0.815
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr1_+_40399618
|
0.810
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr5_-_131160614
|
0.807
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr22_+_38720881
|
0.806
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr4_-_103485600
|
0.804
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr1_-_35957030
|
0.801
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr7_+_154720547
|
0.795
|
|
INSIG1
|
insulin induced gene 1
|
|
chr8_+_95722448
|
0.793
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr8_+_109525050
|
0.786
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr1_+_29086206
|
0.776
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr19_-_52709975
|
0.771
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_+_45250503
|
0.763
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr16_-_66827449
|
0.763
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr7_+_101860958
|
0.763
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr1_+_26609819
|
0.759
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr8_+_38153223
|
0.747
|
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr2_+_198072965
|
0.743
|
NM_002157
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr1_-_115060904
|
0.739
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr8_-_54918389
|
0.739
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr1_-_35956979
|
0.731
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr9_+_130749812
|
0.730
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr2_-_10505902
|
0.727
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr7_+_154720416
|
0.727
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr6_-_33493640
|
0.719
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr19_+_54149941
|
0.719
|
|
BAX
|
BCL2-associated X protein
|
|
chr7_+_101861007
|
0.718
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr8_+_109525010
|
0.715
|
NM_014673
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr18_+_30810889
|
0.714
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_-_18300246
|
0.712
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr5_-_131160541
|
0.705
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr1_+_26609893
|
0.698
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr1_+_29086156
|
0.693
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_+_11788889
|
0.690
|
|
CLCN6
|
chloride channel 6
|
|
chr17_+_54997667
|
0.681
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_-_152459697
|
0.679
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chrX_-_100490612
|
0.676
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_+_205293217
|
0.674
|
NM_001018053
NM_006212
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr1_-_11788550
|
0.674
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr17_-_35273947
|
0.671
|
NM_012481
NM_183228
NM_183229
NM_183230
NM_183231
NM_183232
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr16_+_1341900
|
0.671
|
NM_032520
|
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit
|
|
chr3_+_4996096
|
0.668
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr13_-_45640679
|
0.666
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_10015577
|
0.662
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chrX_+_146870540
|
0.658
|
NM_152578
|
FMR1NB
|
fragile X mental retardation 1 neighbor
|
|
chr11_+_66915997
|
0.656
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr3_-_20202622
|
0.655
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chrX_+_130984912
|
0.650
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr8_-_54918325
|
0.649
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chrX_+_49084499
|
0.638
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE2E
GAGE12J
GAGE2D
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 2E
G antigen 12J
G antigen 2D
G antigen 2A
|
|
chr1_-_26105230
|
0.637
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr22_-_19543099
|
0.633
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr5_+_133735004
|
0.632
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr3_+_50629604
|
0.629
|
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr6_-_43305156
|
0.624
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr10_+_70553936
|
0.622
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrX_-_102452461
|
0.618
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr1_-_52936611
|
0.615
|
NM_023077
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr19_+_6415259
|
0.612
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr8_-_126173203
|
0.611
|
|
KIAA0196
|
KIAA0196
|
|
chr8_+_98725604
|
0.609
|
|
MTDH
|
metadherin
|
|
chr17_+_43373887
|
0.603
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr3_+_4996141
|
0.603
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr7_+_106472243
|
0.603
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chrX_+_134482387
|
0.601
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr10_-_88116181
|
0.600
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr1_+_44213200
|
0.598
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr9_-_125070675
|
0.595
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr9_-_126573349
|
0.591
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr1_-_52936573
|
0.590
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr6_-_31871452
|
0.589
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr19_+_37528340
|
0.588
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr1_-_40335451
|
0.588
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr12_+_110935618
|
0.586
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr1_-_40335496
|
0.581
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chrX_-_107866262
|
0.578
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr9_-_139129015
|
0.578
|
NM_013379
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr7_+_116380590
|
0.576
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chr4_+_101090648
|
0.575
|
|
LOC256880
|
hypothetical LOC256880
|
|
chrX_+_16714452
|
0.574
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr13_+_26896614
|
0.571
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr18_+_9126742
|
0.571
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr9_+_37110506
|
0.570
|
|
ZCCHC7
|
zinc finger, CCHC domain containing 7
|
|
chr4_-_103485419
|
0.569
|
NM_001135147
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr3_+_160002582
|
0.569
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_40335504
|
0.568
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr1_-_224178486
|
0.567
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr12_+_92488932
|
0.566
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr7_+_50314801
|
0.566
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_-_224178475
|
0.565
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_-_224178606
|
0.565
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chrX_-_118583346
|
0.562
|
NM_001170569
|
CXorf56
|
chromosome X open reading frame 56
|
|
chr20_+_60744268
|
0.561
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr5_+_154115046
|
0.560
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr17_+_44325127
|
0.560
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr1_+_45250388
|
0.559
|
NM_000374
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr15_-_81113676
|
0.557
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr8_-_126173055
|
0.556
|
|
KIAA0196
|
KIAA0196
|
|
chr1_-_224178553
|
0.555
|
NM_013328
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr9_+_37110468
|
0.554
|
NM_032226
|
ZCCHC7
|
zinc finger, CCHC domain containing 7
|
|
chr22_-_17799177
|
0.554
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_-_115060857
|
0.554
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr1_-_224178412
|
0.553
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr12_-_47749996
|
0.545
|
NM_144593
|
RHEBL1
|
Ras homolog enriched in brain like 1
|
|
chr5_+_89890366
|
0.544
|
NM_032119
|
GPR98
|
G protein-coupled receptor 98
|
|
chr16_+_22124870
|
0.543
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr9_-_125070613
|
0.540
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr2_-_10505724
|
0.540
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr1_-_28432060
|
0.538
|
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr1_-_227828339
|
0.536
|
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr10_+_22645304
|
0.536
|
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 read-through transcript
COMM domain containing 3
|
|
chr6_-_28999388
|
0.535
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr2_+_177785667
|
0.529
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr15_-_48766213
|
0.527
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_+_51686512
|
0.526
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr18_+_9698187
|
0.525
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_+_133734768
|
0.521
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr1_-_52936602
|
0.516
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr3_-_198509276
|
0.515
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr6_-_28999621
|
0.513
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr6_-_44333002
|
0.513
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr11_-_47620590
|
0.511
|
NM_014342
|
MTCH2
|
mitochondrial carrier homolog 2 (C. elegans)
|
|
chr19_+_55614006
|
0.510
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr9_-_130124317
|
0.510
|
NM_015679
|
TRUB2
|
TruB pseudouridine (psi) synthase homolog 2 (E. coli)
|
|
chr1_+_234625338
|
0.508
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr7_-_8268206
|
0.507
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr19_-_242335
|
0.505
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr10_-_32257693
|
0.505
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr7_-_8268666
|
0.504
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr22_-_17799215
|
0.504
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr4_-_56996400
|
0.500
|
NM_002703
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr8_+_142501188
|
0.500
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr5_-_131160608
|
0.497
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr2_-_10506033
|
0.494
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr1_-_165112093
|
0.492
|
|
TADA1
|
transcriptional adaptor 1
|
|
chr14_-_90953883
|
0.491
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr20_+_60744231
|
0.491
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr12_+_28234657
|
0.489
|
|
|
|
|
chr11_+_107384819
|
0.488
|
|
CUL5
|
cullin 5
|
|
chrX_-_118583401
|
0.488
|
NM_001170570
NM_022101
|
CXorf56
|
chromosome X open reading frame 56
|
|
chr4_-_101090505
|
0.488
|
NM_002106
|
H2AFZ
|
H2A histone family, member Z
|
|
chr12_+_49185035
|
0.487
|
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr8_-_126173189
|
0.486
|
|
KIAA0196
|
KIAA0196
|
|
chr8_+_104496788
|
0.485
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr1_+_44213229
|
0.485
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|