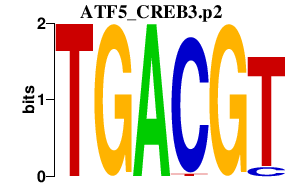
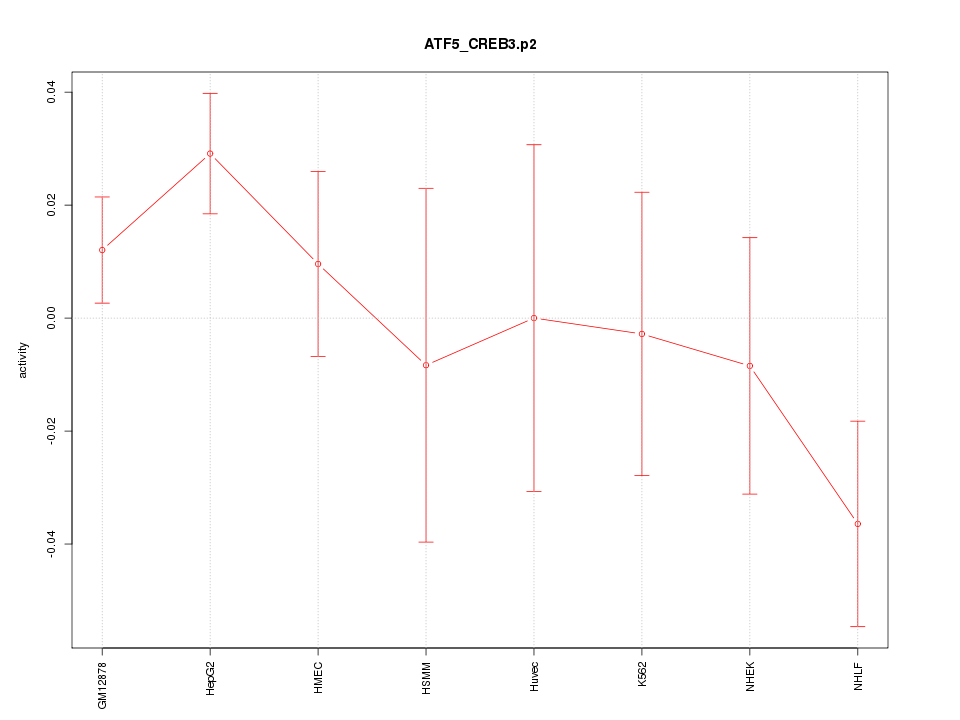
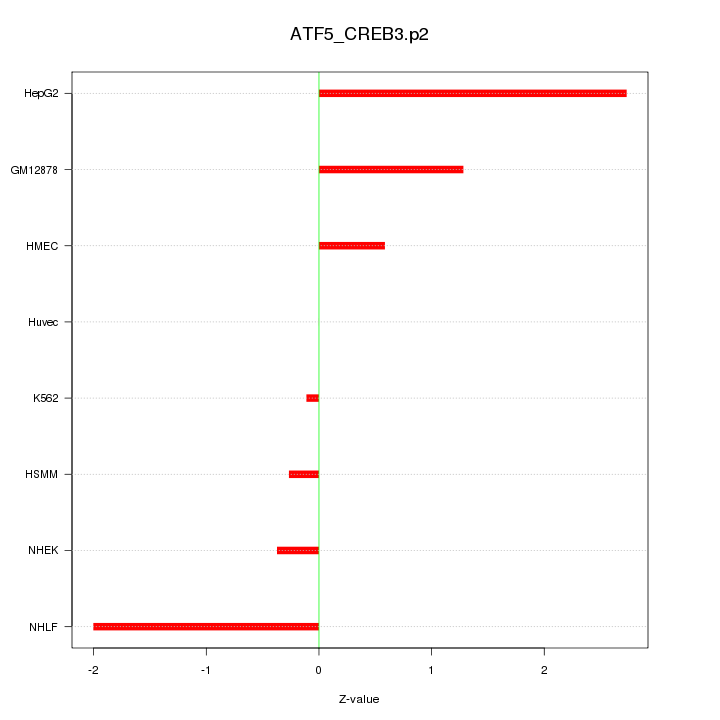
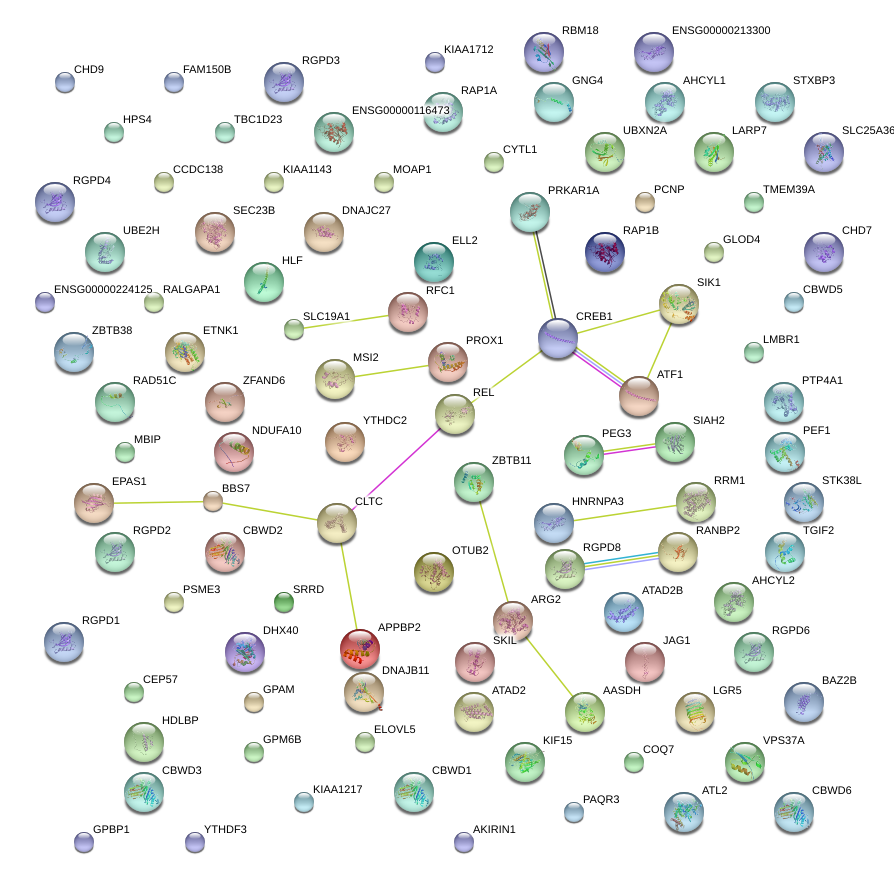
|
chr3_+_171558143
|
2.205
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_101462375
|
1.800
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr3_+_142569988
|
1.727
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr3_+_171558188
|
1.707
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_44778284
|
1.705
|
|
KIF15
|
kinesin family member 15
|
|
chr17_+_52688842
|
1.681
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr3_+_171558215
|
1.654
|
|
SKIL
|
SKI-like oncogene
|
|
chr9_-_69729918
|
1.573
|
|
CBWD5
|
COBW domain containing 5
|
|
chr16_+_18986417
|
1.544
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr9_-_124066901
|
1.430
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr5_+_56505412
|
1.414
|
NM_022913
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr9_-_124066838
|
1.408
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr14_-_35348086
|
1.404
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr17_+_50697355
|
1.373
|
|
HLF
|
hepatic leukemia factor
|
|
chr5_+_56505608
|
1.326
|
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr3_-_102878301
|
1.325
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr17_+_50697319
|
1.317
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr14_+_67156405
|
1.306
|
|
ARG2
|
arginase, type II
|
|
chr14_+_67156386
|
1.300
|
|
ARG2
|
arginase, type II
|
|
chr2_+_208103068
|
1.290
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr3_+_142143329
|
1.288
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr17_-_55958274
|
1.287
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr3_-_44778099
|
1.287
|
NM_020696
|
KIAA1143
|
KIAA1143
|
|
chr3_+_44778212
|
1.259
|
NM_020242
|
KIF15
|
kinesin family member 15
|
|
chr5_-_95323353
|
1.233
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr1_+_110328830
|
1.226
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_-_62043867
|
1.217
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr14_+_67156323
|
1.212
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr17_-_55958073
|
1.202
|
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr2_+_60962206
|
1.201
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr7_-_156378382
|
1.198
|
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr2_+_60962279
|
1.183
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr10_-_113933457
|
1.181
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr2_+_24003648
|
1.177
|
|
UBXN2A
|
UBX domain protein 2A
|
|
chr2_-_240613480
|
1.168
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr4_-_80079393
|
1.165
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr7_-_156378636
|
1.163
|
NM_022458
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr2_-_38457884
|
1.162
|
|
ATL2
|
atlastin GTPase 2
|
|
chr2_-_38457934
|
1.147
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr12_+_22669352
|
1.136
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr12_+_67290881
|
1.116
|
NM_001010942
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr5_+_112877251
|
1.116
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr20_+_18436548
|
1.108
|
NM_001172745
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr6_+_64340408
|
1.105
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_+_208103001
|
1.099
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr12_+_27288475
|
1.098
|
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr12_+_27288446
|
1.092
|
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr20_+_18436525
|
1.091
|
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr5_-_95323180
|
1.083
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr17_+_54997667
|
1.082
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr17_+_64020120
|
1.075
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr2_-_25047988
|
1.057
|
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr1_+_212227902
|
1.055
|
|
PROX1
|
prospero homeobox 1
|
|
chr2_-_24003403
|
1.050
|
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr4_-_39044300
|
1.043
|
NM_002913
|
RFC1
|
replication factor C (activator 1) 1, 145kDa
|
|
chr17_-_632135
|
1.027
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chrX_-_13866565
|
1.026
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr12_+_67290975
|
1.025
|
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr17_+_38238970
|
1.023
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr14_+_93562436
|
1.015
|
NM_023112
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr14_+_93562500
|
1.015
|
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr8_+_17148755
|
1.014
|
NM_001145152
NM_152415
|
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae)
|
|
chr20_+_18436167
|
1.002
|
NM_006363
NM_032985
NM_032986
NM_001172746
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr8_+_64243741
|
0.999
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr7_-_129379947
|
0.995
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr2_-_278181
|
0.991
|
NM_001002919
|
FAM150B
|
family with sequence similarity 150, member B
|
|
chr8_+_64243662
|
0.990
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr17_+_54124952
|
0.990
|
NM_002876
NM_058216
|
RAD51C
|
RAD51 homolog C (S. cerevisiae)
|
|
chr6_+_64339869
|
0.988
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr19_+_47593137
|
0.988
|
|
|
|
|
chr3_-_151963607
|
0.979
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr10_+_24023680
|
0.975
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr1_-_233879617
|
0.966
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr17_+_64020155
|
0.959
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr4_+_113777568
|
0.959
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr6_-_53321545
|
0.949
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr3_+_187771139
|
0.944
|
NM_016306
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr17_+_55051818
|
0.940
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr6_-_53321706
|
0.938
|
NM_021814
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr20_+_34635362
|
0.936
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr4_-_123010918
|
0.933
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr15_+_78139073
|
0.930
|
NM_019006
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr2_-_160181206
|
0.925
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr14_-_35859545
|
0.915
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr14_-_92720935
|
0.914
|
|
MOAP1
|
modulator of apoptosis 1
|
|
chr12_+_22669275
|
0.912
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr21_-_43671355
|
0.909
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr3_+_102775677
|
0.908
|
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr16_+_51722285
|
0.904
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr8_+_61753793
|
0.899
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr11_+_4072635
|
0.897
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr22_-_25209761
|
0.885
|
NM_022081
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr11_+_4072653
|
0.883
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr6_-_53321658
|
0.878
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr17_+_64020159
|
0.870
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr6_+_2190964
|
0.870
|
|
|
|
|
chr17_+_54124928
|
0.867
|
|
RAD51C
|
RAD51 homolog C (S. cerevisiae)
|
|
chr4_+_175441402
|
0.866
|
NM_001040157
|
KIAA1712
|
KIAA1712
|
|
chr22_+_25209812
|
0.857
|
NM_001013694
|
SRRD
|
SRR1 domain containing
|
|
chr1_-_31883051
|
0.855
|
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr2_+_108769572
|
0.853
|
NM_144978
|
CCDC138
|
coiled-coil domain containing 138
|
|
chr2_+_177785667
|
0.852
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr11_+_95163256
|
0.852
|
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr2_-_241860871
|
0.840
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_+_70120028
|
0.839
|
NM_003667
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr3_-_120665113
|
0.833
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr20_-_10602429
|
0.828
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr4_-_56948345
|
0.825
|
NM_181806
|
AASDH
|
aminoadipate-semialdehyde dehydrogenase
|
|
chr2_+_108702338
|
0.823
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr2_-_240613357
|
0.814
|
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr1_+_109090789
|
0.811
|
NM_007269
|
STXBP3
|
syntaxin binding protein 3
|
|
chrX_-_13866366
|
0.810
|
|
|
|
|
chr1_+_39229474
|
0.806
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr17_-_8092078
|
0.806
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr16_+_18986721
|
0.803
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr7_-_23537968
|
0.799
|
NM_013293
|
TRA2A
|
transformer 2 alpha homolog (Drosophila)
|
|
chr17_+_26446067
|
0.798
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr11_+_4072499
|
0.797
|
NM_001033
|
RRM1
|
ribonucleotide reductase M1
|
|
chr19_+_16048306
|
0.795
|
|
TPM4
|
tropomyosin 4
|
|
chr1_+_51855343
|
0.787
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr6_-_122834580
|
0.785
|
NM_020755
|
SERINC1
|
serine incorporator 1
|
|
chr2_+_105320440
|
0.785
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr11_-_111255020
|
0.785
|
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chr17_-_8092098
|
0.784
|
NM_025099
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr3_+_187771192
|
0.784
|
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr14_-_38642165
|
0.779
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr9_+_124066973
|
0.776
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr16_+_20819488
|
0.769
|
NM_001128301
NM_001128302
|
LYRM1
|
LYR motif containing 1
|
|
chr6_+_64340388
|
0.769
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr3_+_38181972
|
0.764
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr1_+_220857774
|
0.763
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr20_+_57948803
|
0.756
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr7_+_96473225
|
0.755
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr12_+_7174230
|
0.754
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr14_-_38971370
|
0.753
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr10_-_59697636
|
0.752
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr12_-_100979869
|
0.752
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr4_-_113777569
|
0.752
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr12_+_67290977
|
0.749
|
|
RAP1B
RAP1BL
|
RAP1B, member of RAS oncogene family
RAP1B, member of RAS oncogene family pseudogene
|
|
chr2_+_208102937
|
0.748
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr17_+_55052046
|
0.746
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr9_+_70046658
|
0.746
|
NM_201453
|
CBWD5
CBWD3
CBWD1
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 1
COBW domain containing 6
|
|
chr1_-_183552833
|
0.745
|
|
IVNS1ABP
|
influenza virus NS1A binding protein
|
|
chr1_+_109090868
|
0.745
|
|
STXBP3
|
syntaxin binding protein 3
|
|
chr1_-_148868644
|
0.745
|
|
ENSA
|
endosulfine alpha
|
|
chrX_+_101910713
|
0.743
|
|
LOC100287765
|
hypothetical LOC100287765
|
|
chr18_+_18767275
|
0.739
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr4_+_153676857
|
0.737
|
|
DKFZP434I0714
|
hypothetical protein DKFZP434I0714
|
|
chrX_+_84385649
|
0.737
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr18_-_257987
|
0.736
|
NM_005131
|
THOC1
|
THO complex 1
|
|
chr14_-_35347791
|
0.732
|
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr3_-_33734747
|
0.729
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr17_-_50854054
|
0.728
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr20_-_42584100
|
0.727
|
NM_006811
NM_198941
|
SERINC3
|
serine incorporator 3
|
|
chr17_+_26060617
|
0.723
|
|
|
|
|
chr6_+_30402599
|
0.723
|
NM_021253
|
TRIM39
|
tripartite motif containing 39
|
|
chr16_+_49139694
|
0.720
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr8_-_117837240
|
0.720
|
NM_003756
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H
|
|
chrX_+_24077782
|
0.719
|
|
ZFX
|
zinc finger protein, X-linked
|
|
chr8_-_17148637
|
0.719
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr1_+_169721260
|
0.718
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chrX_-_13745134
|
0.715
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_72897323
|
0.713
|
NM_032175
|
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae)
|
|
chr10_-_97406357
|
0.711
|
|
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr1_-_148868713
|
0.710
|
NM_004436
NM_207042
NM_207043
NM_207044
NM_207168
|
ENSA
|
endosulfine alpha
|
|
chr12_+_81276406
|
0.708
|
NM_032230
|
C12orf26
|
chromosome 12 open reading frame 26
|
|
chr8_+_123863224
|
0.702
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_+_16048266
|
0.699
|
|
TPM4
|
tropomyosin 4
|
|
chr2_+_183289205
|
0.696
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr3_-_197293377
|
0.693
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chrX_+_24077649
|
0.692
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chrX_-_54086318
|
0.689
|
NM_001184898
|
PHF8
|
PHD finger protein 8
|
|
chr4_-_186584031
|
0.689
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr5_+_139534812
|
0.688
|
NM_032412
|
C5orf32
|
chromosome 5 open reading frame 32
|
|
chr17_-_46553083
|
0.687
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_+_170778265
|
0.686
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr11_-_8636958
|
0.686
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr8_+_110621092
|
0.685
|
NM_198120
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9
|
|
chr12_+_27288344
|
0.685
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr11_+_113776582
|
0.685
|
NM_016090
|
RBM7
|
RNA binding motif protein 7
|
|
chr2_+_191453813
|
0.684
|
|
GLS
|
glutaminase
|
|
chr3_+_38182120
|
0.684
|
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr3_+_16281670
|
0.683
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr2_-_86704446
|
0.682
|
|
RNF103
|
ring finger protein 103
|
|
chr4_+_85723080
|
0.680
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr2_+_170149092
|
0.679
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr3_-_33734544
|
0.678
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_186786655
|
0.677
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr8_-_103493637
|
0.676
|
NM_015902
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5
|
|
chr2_+_74610751
|
0.676
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr17_+_55052060
|
0.676
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr4_+_113778060
|
0.673
|
NM_015454
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr14_+_44501083
|
0.672
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr5_-_72896964
|
0.671
|
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr14_-_38642029
|
0.671
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr17_+_38238942
|
0.668
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr1_+_110328955
|
0.667
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_-_183553083
|
0.666
|
NM_006469
|
IVNS1ABP
|
influenza virus NS1A binding protein
|
|
chr8_-_17148474
|
0.663
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr10_-_115923874
|
0.661
|
|
C10orf118
|
chromosome 10 open reading frame 118
|