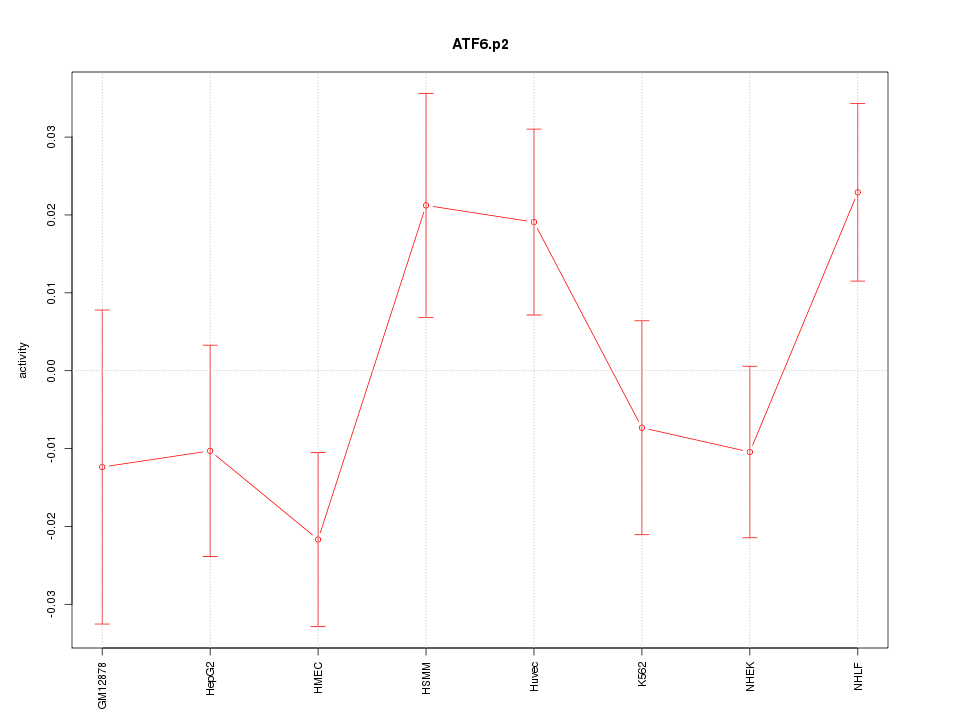
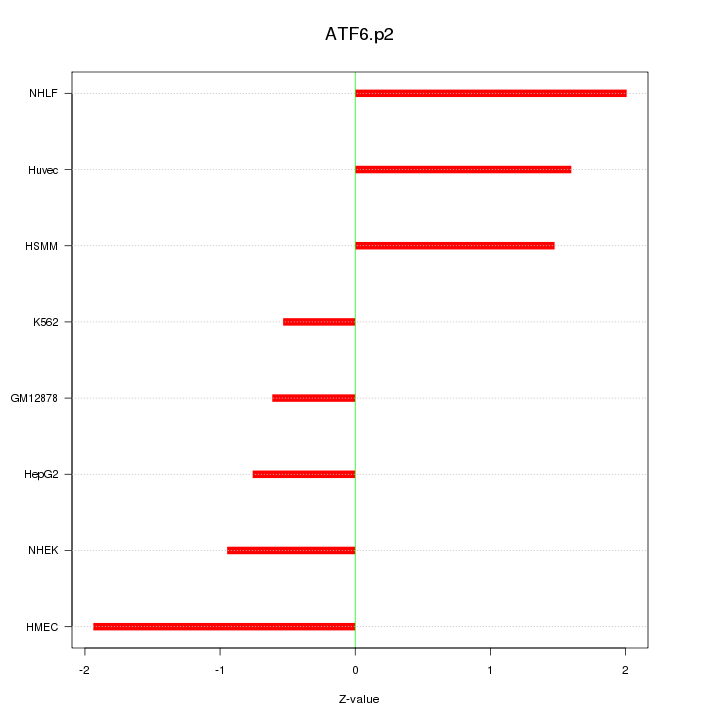
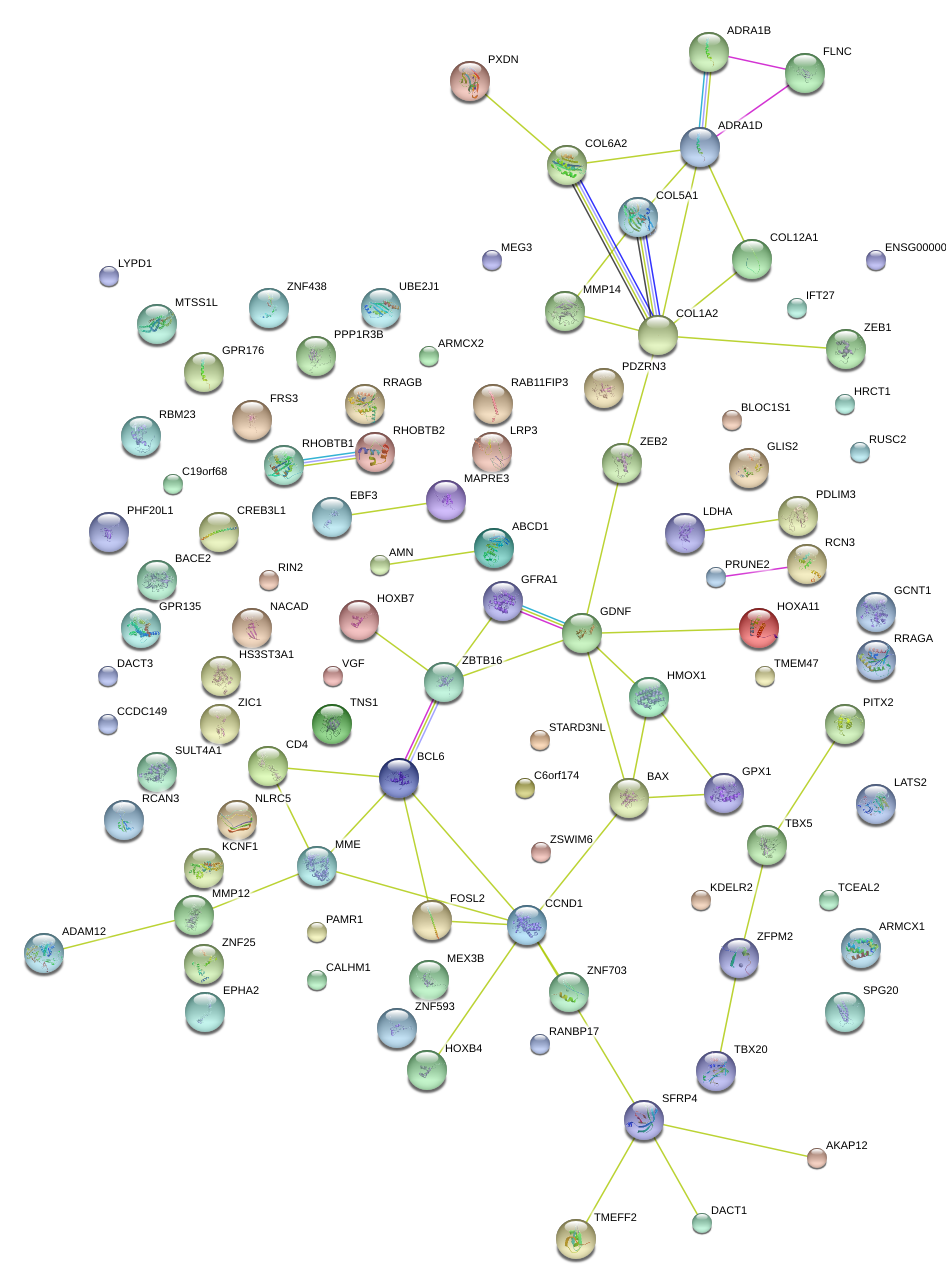
|
chr17_-_44043361
|
1.776
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chrX_-_100801472
|
1.631
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr7_+_128257698
|
1.596
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr1_+_26368969
|
1.342
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr2_+_28469172
|
1.144
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr14_+_100362204
|
1.067
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr17_-_44042924
|
1.061
|
|
HOXB7
|
homeobox B7
|
|
chr7_+_27191534
|
0.897
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr15_-_80125403
|
0.883
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr4_-_186693574
|
0.865
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr12_+_110935612
|
0.840
|
|
|
|
|
chr2_+_28469275
|
0.814
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr19_+_54723044
|
0.784
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr15_-_80125514
|
0.783
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr9_-_78710698
|
0.777
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr7_-_27191287
|
0.777
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr7_+_93861808
|
0.754
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr17_-_44043278
|
0.727
|
|
HOXB7
|
homeobox B7
|
|
chr10_-_62373828
|
0.720
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr5_+_170221584
|
0.701
|
NM_022897
|
RANBP17
|
RAN binding protein 17
|
|
chr12_+_54396086
|
0.688
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr17_-_13445942
|
0.668
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr10_-_118022965
|
0.656
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr10_-_128067062
|
0.653
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr3_-_188945921
|
0.639
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_-_73756668
|
0.596
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr7_-_35260213
|
0.587
|
NM_001077653
NM_001166220
|
TBX20
|
T-box 20
|
|
chr12_-_113328271
|
0.582
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr8_+_106400322
|
0.577
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chrX_-_34585287
|
0.575
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr2_-_144994038
|
0.564
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_+_100692071
|
0.558
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr3_-_188946168
|
0.557
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr15_-_38000379
|
0.554
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr10_-_131652364
|
0.547
|
|
EBF3
|
early B-cell factor 3
|
|
chr2_+_28469199
|
0.531
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_-_111763655
|
0.527
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr9_+_35528890
|
0.524
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr6_-_75972473
|
0.520
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr12_+_6768960
|
0.518
|
|
CD4
|
CD4 molecule
|
|
chr2_-_1727292
|
0.508
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr14_+_102458745
|
0.502
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr2_-_192767494
|
0.497
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr14_+_100362236
|
0.496
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_-_35503720
|
0.494
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr9_+_136673464
|
0.489
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr13_-_20533629
|
0.487
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr7_-_6490375
|
0.486
|
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr20_+_19863716
|
0.468
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr2_-_1727247
|
0.467
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr9_+_35528628
|
0.467
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_+_10969513
|
0.466
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr22_-_42589543
|
0.465
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr10_-_128067001
|
0.465
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr16_-_69277355
|
0.459
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr3_+_148609841
|
0.452
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr13_-_35818396
|
0.442
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr16_+_4325142
|
0.441
|
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr12_+_6768934
|
0.439
|
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr5_-_37875538
|
0.431
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr20_-_4177519
|
0.431
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr17_-_44010543
|
0.428
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr2_-_133144094
|
0.426
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr11_+_113435506
|
0.423
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr13_-_35818777
|
0.418
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr14_+_22375632
|
0.418
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chrX_+_55760774
|
0.415
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr14_-_59001804
|
0.409
|
NM_022571
|
GPR135
|
G protein-coupled receptor 135
|
|
chr6_+_135860631
|
0.406
|
|
NCRNA00271
|
non-protein coding RNA 271
|
|
chr8_+_133856738
|
0.406
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr13_-_35818603
|
0.404
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr1_-_16355013
|
0.402
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr10_-_105208634
|
0.393
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr7_-_37923040
|
0.391
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr5_+_159276317
|
0.387
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr8_+_37672427
|
0.386
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr19_+_54149843
|
0.378
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr22_-_35502059
|
0.373
|
NM_001177701
NM_006860
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr6_-_90119068
|
0.373
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr14_+_22375747
|
0.372
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr10_-_31360778
|
0.371
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr21_+_46369606
|
0.368
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr16_+_55580910
|
0.368
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr11_+_69165119
|
0.367
|
|
CCND1
|
cyclin D1
|
|
chr11_+_69165065
|
0.367
|
|
CCND1
|
cyclin D1
|
|
chr12_+_6768966
|
0.366
|
|
CD4
|
CD4 molecule
|
|
chr19_+_38376980
|
0.366
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr7_-_45095017
|
0.365
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr12_+_6768866
|
0.363
|
NM_000616
|
CD4
|
CD4 molecule
|
|
chr3_-_49370755
|
0.362
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr6_+_151602787
|
0.362
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chrX_+_101267315
|
0.360
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr11_+_69165018
|
0.358
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr19_-_55046117
|
0.356
|
|
LOC100506033
|
hypothetical LOC100506033
|
|
chr6_-_127822227
|
0.356
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr11_+_113435640
|
0.355
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr11_+_46255737
|
0.354
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr14_-_22458134
|
0.351
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr14_+_22375753
|
0.350
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr16_+_415606
|
0.349
|
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr5_+_60663842
|
0.347
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr14_+_22375656
|
0.346
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr3_-_49370682
|
0.345
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr2_-_218575922
|
0.344
|
|
TNS1
|
tensin 1
|
|
chr7_-_100595552
|
0.340
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr22_+_34107087
|
0.337
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr3_+_156280772
|
0.336
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr14_+_58174489
|
0.335
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr8_-_9046460
|
0.334
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr11_+_18372682
|
0.331
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr7_+_38184393
|
0.329
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr19_+_53365748
|
0.327
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr4_-_24523605
|
0.327
|
NM_001130726
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr9_+_35896188
|
0.326
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr21_+_41461597
|
0.324
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr9_+_78263887
|
0.324
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr10_-_38305458
|
0.323
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr7_+_38184444
|
0.323
|
NM_032016
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr2_-_216008689
|
0.321
|
|
FN1
|
fibronectin 1
|
|
chr11_+_18372687
|
0.321
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr4_-_47160476
|
0.320
|
|
COMMD8
|
COMM domain containing 8
|
|
chr9_-_103289154
|
0.319
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr3_+_50272648
|
0.318
|
|
|
|
|
chr7_+_5598964
|
0.318
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr21_-_37367259
|
0.317
|
NM_153682
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr1_-_206484258
|
0.314
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr7_+_12692938
|
0.314
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr10_+_102749597
|
0.314
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr18_+_54682261
|
0.312
|
|
ZNF532
|
zinc finger protein 532
|
|
chr6_+_1336077
|
0.312
|
|
|
|
|
chr17_+_56884560
|
0.311
|
|
TBX4
|
T-box 4
|
|
chr10_-_128067052
|
0.311
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr22_+_34107051
|
0.311
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr11_+_18372689
|
0.309
|
|
LDHA
|
lactate dehydrogenase A
|
|
chrX_+_47326594
|
0.309
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr7_+_93862137
|
0.309
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr17_-_76064842
|
0.308
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr19_+_18655391
|
0.308
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr14_+_23905984
|
0.307
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr19_+_18655483
|
0.306
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr1_+_219119358
|
0.306
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr8_-_95343716
|
0.305
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_-_25328452
|
0.304
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr14_-_60260211
|
0.302
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr5_+_53849251
|
0.302
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr4_-_141293572
|
0.300
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr9_-_72218098
|
0.298
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr13_-_101852028
|
0.293
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr13_-_44048391
|
0.292
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr16_+_65840275
|
0.291
|
NM_004594
|
SLC9A5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr5_-_172130766
|
0.290
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr15_+_41597097
|
0.289
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr22_-_29833484
|
0.286
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr8_-_18585721
|
0.284
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr8_-_82186832
|
0.283
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr17_-_45829863
|
0.283
|
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr7_-_72822471
|
0.282
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr2_-_216009015
|
0.280
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr11_+_119701047
|
0.280
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr5_-_95794416
|
0.279
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr1_-_201420927
|
0.279
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr19_+_50041400
|
0.278
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr11_+_18372708
|
0.277
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr10_+_70553993
|
0.277
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr7_+_38184375
|
0.277
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chrX_+_134306337
|
0.277
|
NM_152695
|
ZNF449
|
zinc finger protein 449
|
|
chr20_+_34523231
|
0.276
|
NM_001042486
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr2_-_1727324
|
0.276
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr12_+_70434920
|
0.276
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr6_-_90119158
|
0.271
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr7_-_19123787
|
0.270
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr4_+_8645421
|
0.269
|
|
CPZ
|
carboxypeptidase Z
|
|
chr17_-_37828643
|
0.269
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr1_-_181654124
|
0.266
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr5_+_133734768
|
0.265
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr1_+_181871830
|
0.265
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr3_+_50259329
|
0.263
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr7_-_6490214
|
0.263
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr6_-_90119008
|
0.263
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr6_-_75972286
|
0.261
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr2_-_236741352
|
0.260
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr7_-_19123765
|
0.259
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr6_-_91063181
|
0.257
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr19_+_50041424
|
0.257
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr14_-_74713062
|
0.257
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chrX_-_83644054
|
0.256
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chrX_-_128485121
|
0.254
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr4_-_114901789
|
0.253
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr12_+_64869271
|
0.253
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr19_+_43502591
|
0.252
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr3_+_127118130
|
0.251
|
|
LOC100125556
|
family with sequence similarity 86, member A pseudogene
|
|
chr10_+_119291945
|
0.251
|
NM_001165924
NM_004098
|
EMX2
|
empty spiracles homeobox 2
|
|
chr15_-_40052041
|
0.251
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr10_+_112247614
|
0.250
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr7_-_37922902
|
0.250
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr19_-_41397396
|
0.249
|
NM_152477
|
ZNF565
|
zinc finger protein 565
|
|
chr15_-_53668341
|
0.248
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr19_+_43502380
|
0.247
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr5_+_170221441
|
0.246
|
|
RANBP17
|
RAN binding protein 17
|
|
chr2_-_73193566
|
0.246
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|