|
chr4_+_39874921
|
2.261
|
NM_004310
|
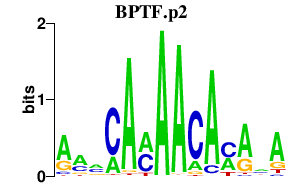
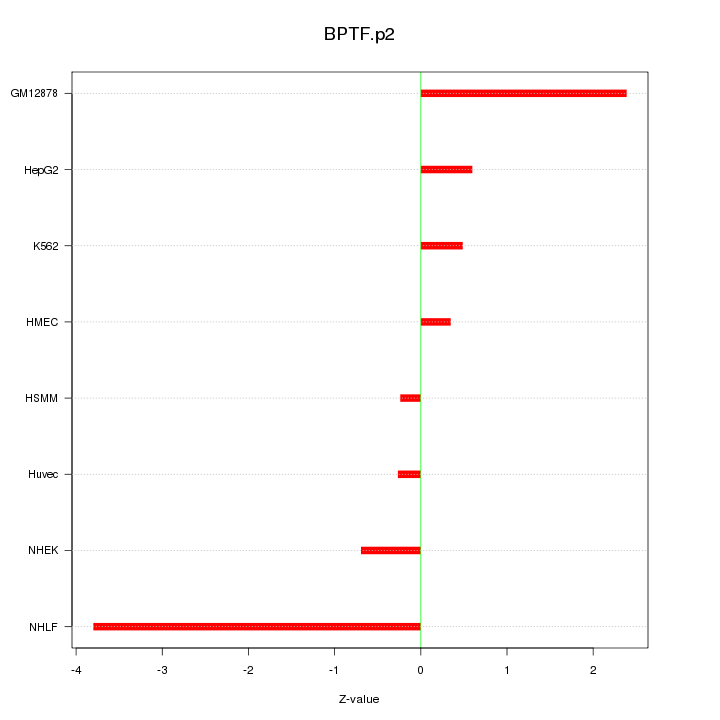
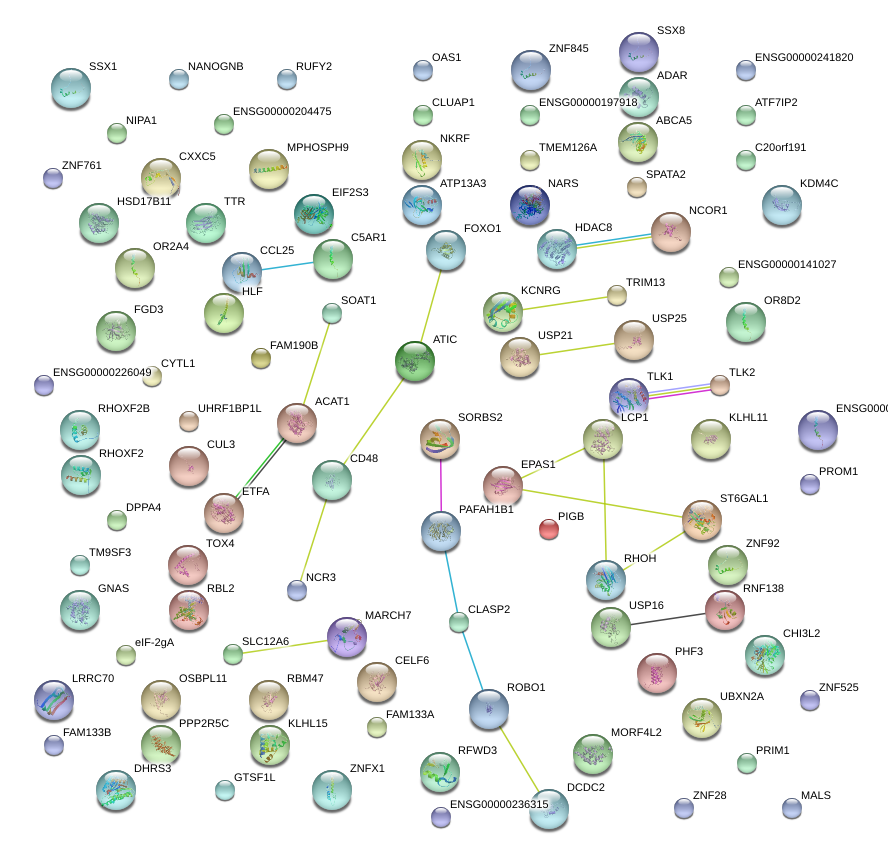
RHOH
|
ras homolog gene family, member H
|
|
chr19_+_52504914
|
1.328
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr2_-_225073357
|
1.261
|
|
CUL3
|
cullin 3
|
|
chrX_-_71709622
|
1.256
|
NM_001166418
NM_001166419
NM_001166420
NM_001166422
NM_001166448
NM_018486
|
HDAC8
|
histone deacetylase 8
|
|
chr18_+_27425727
|
1.223
|
NM_000371
|
TTR
|
transthyretin
|
|
chr3_+_188222337
|
1.206
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chrX_+_119176494
|
1.190
|
NM_001099685
NM_032498
|
RHOXF2B
RHOXF2
|
Rhox homeobox family, member 2B
Rhox homeobox family, member 2
|
|
chr4_-_40212670
|
1.145
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr12_+_7809078
|
1.136
|
NM_001145465
|
NANOGNB
|
NANOG neighbor homeobox
|
|
chr1_-_12599934
|
1.120
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr2_+_215884923
|
1.095
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr21_+_29322100
|
1.008
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr17_-_59818033
|
0.971
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr13_-_45654327
|
0.958
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_45654292
|
0.958
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_+_10387412
|
0.956
|
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr4_-_88531353
|
0.937
|
NM_016245
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr9_+_6710862
|
0.933
|
NM_001146696
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr5_-_42847716
|
0.925
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr10_-_98311813
|
0.918
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chrX_-_23955223
|
0.906
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr2_+_215885053
|
0.898
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr15_-_74390861
|
0.884
|
NM_000126
NM_001127716
|
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide
|
|
chr3_-_33734544
|
0.865
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr13_-_45654296
|
0.863
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_+_52037186
|
0.854
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr10_-_69836912
|
0.847
|
NM_017987
NM_001042417
|
RUFY2
|
RUN and FYVE domain containing 2
|
|
chr3_-_110539023
|
0.835
|
|
DPPA4
|
developmental pluripotency associated 4
|
|
chrX_-_118610880
|
0.832
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr15_+_53398424
|
0.827
|
NM_004855
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr1_+_111571803
|
0.819
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr2_+_24016771
|
0.810
|
NM_181713
|
UBXN2A
|
UBX domain protein 2A
|
|
chr12_+_111829191
|
0.797
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr17_-_1195096
|
0.786
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr16_+_3499894
|
0.781
|
NM_024793
|
CLUAP1
|
clusterin associated protein 1
|
|
chr15_+_53398697
|
0.761
|
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr9_+_94749421
|
0.742
|
NM_001083536
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr16_-_73258276
|
0.733
|
NM_018124
|
RFWD3
|
ring finger and WD repeat domain 3
|
|
chr19_-_58016696
|
0.728
|
NM_006969
|
ZNF28
|
zinc finger protein 28
|
|
chr3_-_126797057
|
0.724
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr13_-_40138607
|
0.715
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr7_+_64476167
|
0.714
|
NM_007139
NM_152626
|
ZNF92
|
zinc finger protein 92
|
|
chr6_+_64414389
|
0.714
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr17_-_64823368
|
0.708
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr7_+_64476226
|
0.702
|
|
ZNF92
|
zinc finger protein 92
|
|
chr1_-_158948176
|
0.698
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr2_-_171631728
|
0.682
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr19_+_8023887
|
0.678
|
NM_005624
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr5_+_139008091
|
0.674
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr18_+_27925923
|
0.671
|
|
RNF138
|
ring finger protein 138
|
|
chr12_-_55432348
|
0.661
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr17_-_37275142
|
0.659
|
NM_018143
|
KLHL11
|
kelch-like 11 (Drosophila)
|
|
chr6_-_24466258
|
0.657
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr20_-_41788975
|
0.636
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr15_-_20638283
|
0.635
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr3_-_195670256
|
0.635
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr15_-_70385709
|
0.634
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr6_-_31668476
|
0.631
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr7_-_143587653
|
0.628
|
NM_001005328
|
OR2A7
OR2A4
|
olfactory receptor, family 2, subfamily A, member 7
olfactory receptor, family 2, subfamily A, member 4
|
|
chr11_+_107497462
|
0.627
|
NM_000019
|
ACAT1
|
acetyl-CoA acetyltransferase 1
|
|
chr2_+_160277273
|
0.626
|
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr4_-_88531245
|
0.625
|
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chrX_+_23982992
|
0.624
|
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa
|
|
chr12_-_122272346
|
0.610
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr4_-_187114863
|
0.608
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_-_53440005
|
0.603
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr16_-_73258248
|
0.602
|
|
RFWD3
|
ring finger and WD repeat domain 3
|
|
chr18_+_27925815
|
0.592
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr12_-_99060771
|
0.591
|
NM_001006947
NM_015054
|
UHRF1BP1L
|
UHRF1 binding protein 1-like
|
|
chrX_+_47999740
|
0.589
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr17_+_50697319
|
0.586
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr3_-_79899748
|
0.584
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr13_+_49487390
|
0.583
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chrX_-_102829621
|
0.582
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr20_-_47307554
|
0.581
|
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr5_+_61910318
|
0.579
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr17_-_15914485
|
0.579
|
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr20_+_56903595
|
0.576
|
|
GNAS
|
GNAS complex locus
|
|
chr15_+_39363442
|
0.575
|
|
LOC729082
|
hypothetical LOC729082
|
|
chr15_-_32398220
|
0.574
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_-_152847256
|
0.570
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr20_-_47963682
|
0.567
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr4_-_15694764
|
0.565
|
|
PROM1
|
prominin 1
|
|
chr14_+_101345878
|
0.565
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr7_-_92057597
|
0.559
|
NM_001040057
NM_152789
|
FAM133B
|
family with sequence similarity 133, member B
|
|
chr10_+_86078136
|
0.557
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr9_+_127002630
|
0.556
|
NM_001174152
NM_001174153
|
RABEPK
|
Rab9 effector protein with kelch motifs
|
|
chr3_+_19963534
|
0.555
|
NM_004162
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chrX_+_23982981
|
0.555
|
NM_001415
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa
|
|
chr15_-_60139857
|
0.552
|
NM_001018088
NM_017684
NM_018080
NM_020821
|
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae)
|
|
chrX_-_80344096
|
0.550
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chrX_-_19443299
|
0.549
|
NM_001001671
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15
|
|
chr1_+_26744893
|
0.548
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr8_+_42368540
|
0.547
|
|
VDAC3
|
voltage-dependent anion channel 3
|
|
chr22_+_17012841
|
0.547
|
|
USP18
|
ubiquitin specific peptidase 18
|
|
chr16_+_24648446
|
0.528
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr13_-_98757634
|
0.527
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr1_-_93418833
|
0.526
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr12_-_99060590
|
0.525
|
|
UHRF1BP1L
|
UHRF1 binding protein 1-like
|
|
chr22_+_35290271
|
0.522
|
|
|
|
|
chr10_-_32707549
|
0.521
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr8_-_37847095
|
0.521
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr1_+_46910086
|
0.521
|
NM_001145474
|
C1orf223
|
chromosome 1 open reading frame 223
|
|
chrX_-_76824726
|
0.517
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr3_-_161584137
|
0.517
|
NM_001190242
|
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas)
|
|
chr2_+_201464884
|
0.514
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr6_+_42857540
|
0.509
|
|
KIAA0240
|
KIAA0240
|
|
chr10_-_93992982
|
0.504
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr15_-_74390846
|
0.503
|
|
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide
|
|
chr15_+_57184568
|
0.503
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr2_+_61243132
|
0.502
|
NM_001143959
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr3_-_33734747
|
0.496
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_142175329
|
0.496
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr22_+_21079355
|
0.495
|
|
|
|
|
chr5_+_139856688
|
0.495
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chr22_+_38652555
|
0.494
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr4_+_170817787
|
0.493
|
|
CLCN3
|
chloride channel 3
|
|
chr12_-_54607925
|
0.493
|
|
WIBG
|
within bgcn homolog (Drosophila)
|
|
chr16_-_73258218
|
0.489
|
|
RFWD3
|
ring finger and WD repeat domain 3
|
|
chr12_+_73219826
|
0.489
|
|
|
|
|
chr17_+_50697355
|
0.488
|
|
HLF
|
hepatic leukemia factor
|
|
chr12_-_93921586
|
0.486
|
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr2_+_3600784
|
0.485
|
|
RPS7
|
ribosomal protein S7
|
|
chrX_+_41433169
|
0.484
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr9_+_5646582
|
0.484
|
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chrX_-_12972641
|
0.484
|
NM_174901
|
FAM9C
|
family with sequence similarity 9, member C
|
|
chr3_-_52688768
|
0.483
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr2_+_215885063
|
0.480
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr2_+_108769572
|
0.480
|
NM_144978
|
CCDC138
|
coiled-coil domain containing 138
|
|
chr4_+_53986759
|
0.479
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr11_-_47620590
|
0.479
|
NM_014342
|
MTCH2
|
mitochondrial carrier homolog 2 (C. elegans)
|
|
chr11_-_3759886
|
0.477
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr10_+_82158186
|
0.477
|
NM_032333
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr7_+_74962234
|
0.475
|
NM_001099435
|
SPDYE5
|
speedy homolog E5 (Xenopus laevis)
|
|
chrX_-_119486874
|
0.475
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr11_-_33847834
|
0.473
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_-_39199936
|
0.469
|
|
|
|
|
chr6_-_24491498
|
0.468
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr11_-_77411933
|
0.467
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr16_+_28850760
|
0.467
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr1_+_39569396
|
0.466
|
NM_033044
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr13_-_24395054
|
0.466
|
NM_018451
|
CENPJ
|
centromere protein J
|
|
chr2_-_44440358
|
0.465
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr6_+_109557199
|
0.463
|
NM_173830
|
CEP57L1
|
centrosomal protein 57kDa-like 1
|
|
chrX_-_19815370
|
0.460
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_-_100979869
|
0.459
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr17_-_34860840
|
0.455
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr13_-_40735619
|
0.454
|
NM_004294
|
MTRF1
|
mitochondrial translational release factor 1
|
|
chr7_-_149960218
|
0.452
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr17_+_34871214
|
0.451
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr17_-_57295451
|
0.447
|
NM_032043
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr3_+_173240107
|
0.447
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_+_78017894
|
0.446
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr4_-_21308415
|
0.445
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr21_-_39739528
|
0.443
|
|
LCA5L
|
Leber congenital amaurosis 5-like
|
|
chr5_-_77841686
|
0.442
|
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr20_+_56904042
|
0.442
|
|
GNAS
|
GNAS complex locus
|
|
chr15_-_76378957
|
0.441
|
NM_025234
|
WDR61
|
WD repeat domain 61
|
|
chr11_-_77026406
|
0.439
|
NM_001293
|
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr19_-_44014228
|
0.438
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr2_+_85658235
|
0.437
|
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chr1_-_218329773
|
0.437
|
NM_006085
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chrX_-_70243362
|
0.436
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr1_-_165789640
|
0.435
|
NM_003851
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr2_-_232034638
|
0.435
|
|
NCL
|
nucleolin
|
|
chr13_-_24394913
|
0.434
|
|
CENPJ
|
centromere protein J
|
|
chr20_-_33580861
|
0.433
|
NM_001145350
|
C20orf173
|
chromosome 20 open reading frame 173
|
|
chr6_-_24827265
|
0.432
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr1_+_221955876
|
0.432
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_144595224
|
0.432
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr6_+_44295219
|
0.432
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr15_-_41904059
|
0.431
|
NM_005926
|
MFAP1
|
microfibrillar-associated protein 1
|
|
chr12_+_111375382
|
0.429
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr9_-_98580148
|
0.429
|
NM_014930
|
ZNF510
|
zinc finger protein 510
|
|
chr16_+_8714326
|
0.428
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_-_12182641
|
0.428
|
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chrX_+_24393264
|
0.427
|
NM_001142386
NM_005391
|
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3
|
|
chr19_+_23737651
|
0.427
|
|
RPSAP58
|
ribosomal protein SA pseudogene 58
|
|
chr12_-_44919818
|
0.426
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr19_+_39402167
|
0.425
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr1_-_165789586
|
0.425
|
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr10_-_29963847
|
0.425
|
|
|
|
|
chr15_-_74390797
|
0.424
|
|
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide
|
|
chr18_-_53440012
|
0.423
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr5_-_32439067
|
0.423
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr16_-_57325633
|
0.422
|
NM_002080
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr1_-_204852465
|
0.421
|
NM_006893
|
EIF2D
|
eukaryotic translation initiation factor 2D
|
|
chrX_-_15242589
|
0.420
|
NM_001012428
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chrX_-_32083506
|
0.420
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr16_+_4485859
|
0.419
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr5_+_161427225
|
0.418
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr12_+_32723539
|
0.418
|
|
DNM1L
|
dynamin 1-like
|
|
chr6_+_44418374
|
0.418
|
NM_145026
|
SPATS1
|
spermatogenesis associated, serine-rich 1
|
|
chr11_+_120966280
|
0.417
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr6_-_149847718
|
0.416
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr4_-_15694691
|
0.416
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr14_+_38804226
|
0.415
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr13_+_77213295
|
0.415
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr1_-_165789630
|
0.415
|
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr15_-_53398534
|
0.414
|
|
|
|