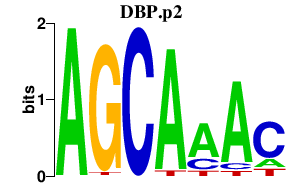
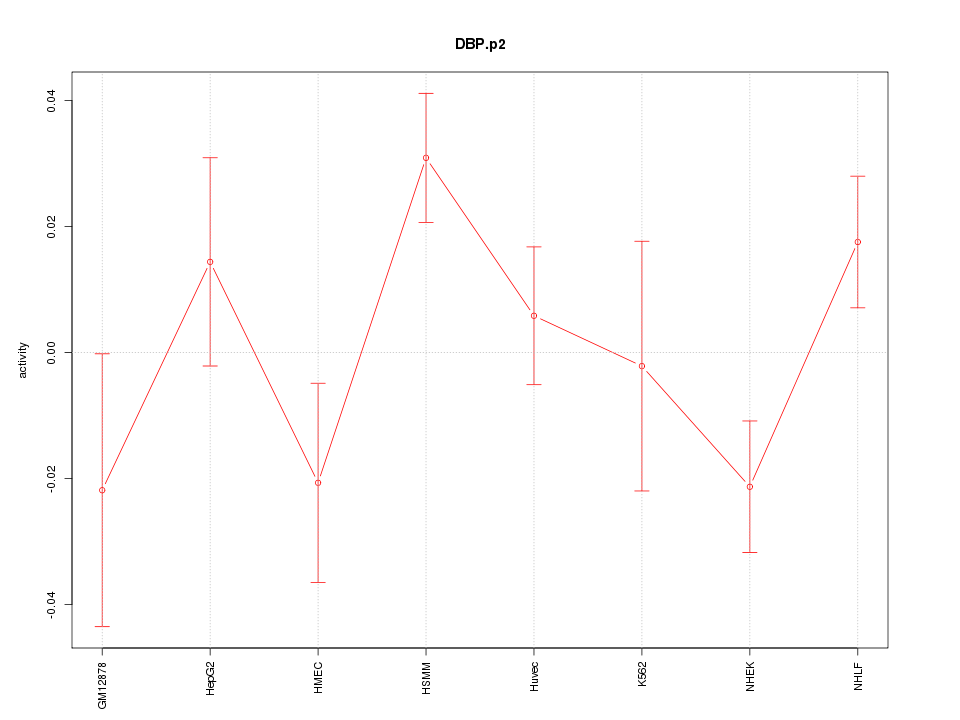
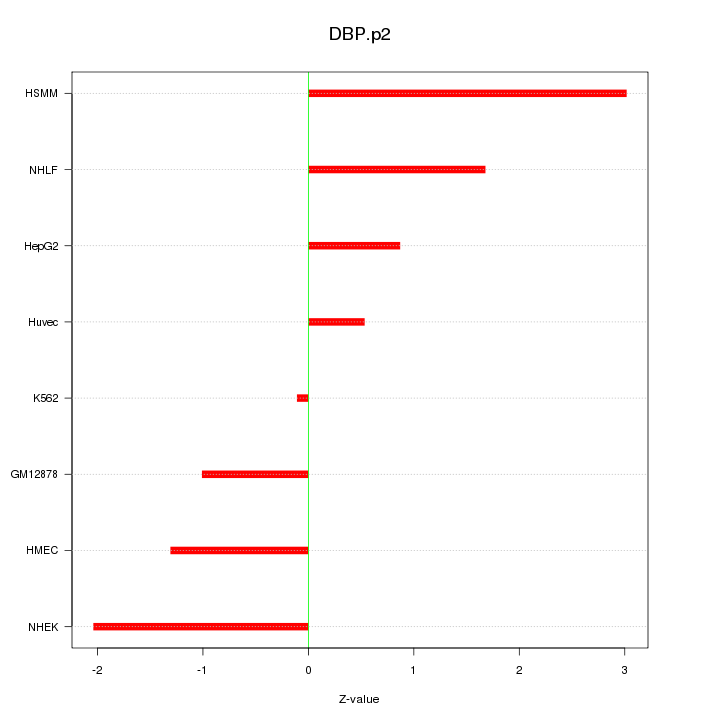
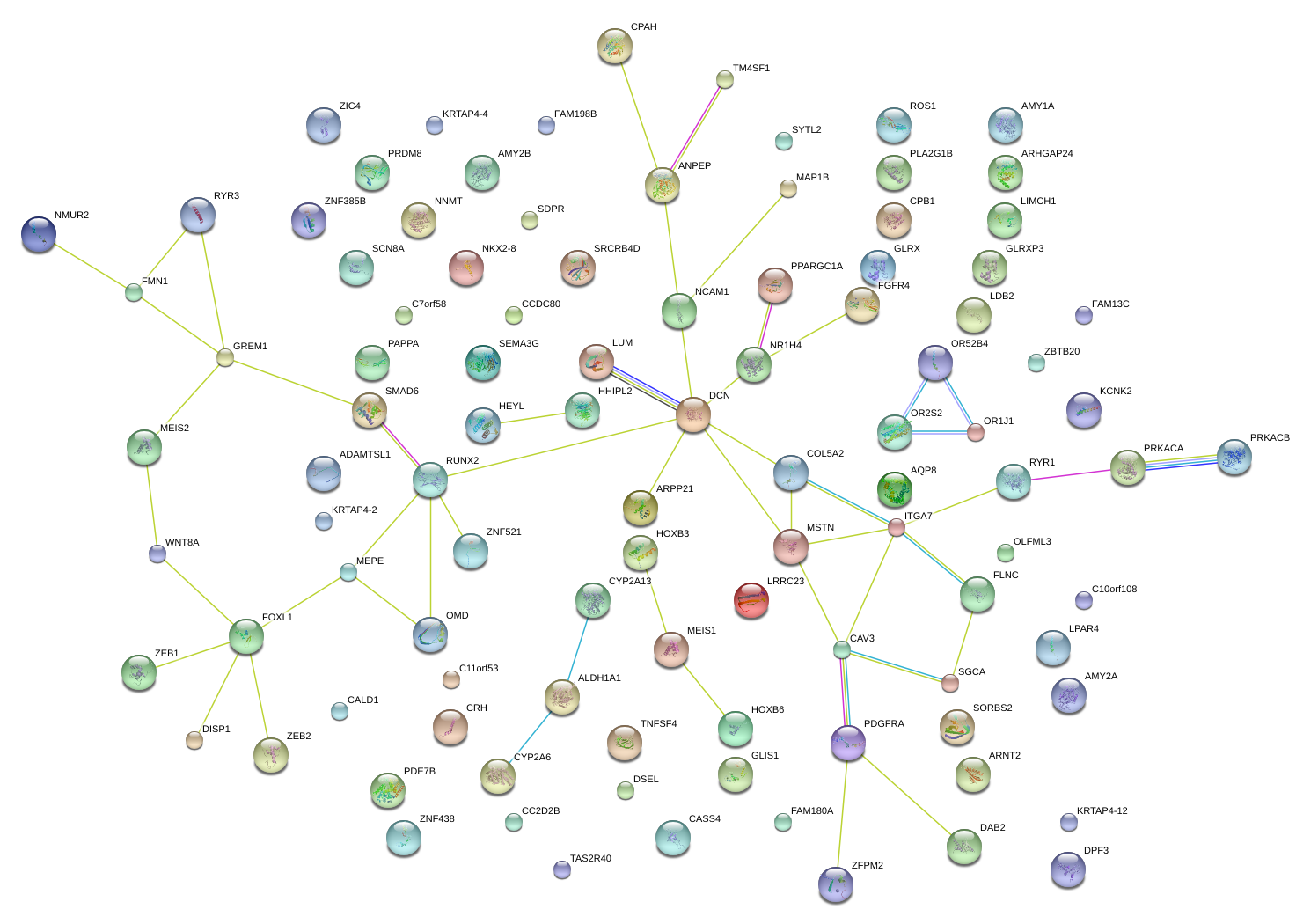
|
chr12_+_50342814
|
1.218
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr2_-_189752575
|
1.176
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr14_-_72430545
|
1.136
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr1_+_114323552
|
1.112
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr7_+_120416680
|
1.102
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr17_-_44006808
|
1.093
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr4_-_159313613
|
1.074
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr15_-_88150823
|
1.071
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr1_+_221168388
|
1.047
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr4_-_186969202
|
1.042
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186969041
|
1.000
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_53972464
|
0.982
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr18_-_21184824
|
0.969
|
|
|
|
|
chr1_+_84382529
|
0.964
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_-_36533903
|
0.959
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr1_-_171442937
|
0.944
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr12_-_90029328
|
0.931
|
|
LUM
|
lumican
|
|
chr12_-_90029672
|
0.918
|
NM_002345
|
LUM
|
lumican
|
|
chr6_+_45404031
|
0.914
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr11_-_85107569
|
0.906
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_+_18464090
|
0.901
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr3_+_8750485
|
0.882
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr5_-_151764857
|
0.862
|
NM_020167
|
NMUR2
|
neuromedin U receptor 2
|
|
chr4_-_23500706
|
0.859
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr10_+_685887
|
0.859
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr10_+_97749837
|
0.844
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr2_-_192419896
|
0.843
|
|
SDPR
|
serum deprivation response
|
|
chr3_-_148604760
|
0.838
|
NM_001168378
|
ZIC4
|
Zic family member 4
|
|
chr12_+_99391681
|
0.813
|
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr6_-_117853710
|
0.809
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr4_-_159312973
|
0.807
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr18_-_63333487
|
0.805
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr11_+_112337411
|
0.804
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr1_+_103961476
|
0.792
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr17_-_44037332
|
0.789
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chrX_+_77889861
|
0.783
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr8_-_68821131
|
0.780
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr5_+_71538456
|
0.772
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr18_-_21185968
|
0.761
|
|
ZNF521
|
zinc finger protein 521
|
|
chr11_-_4346191
|
0.745
|
NM_001005161
|
OR52B4
|
olfactory receptor, family 52, subfamily B, member 4
|
|
chr3_-_150578023
|
0.732
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr4_+_87070449
|
0.727
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_-_94226380
|
0.720
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr15_-_31147376
|
0.719
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr16_+_85169615
|
0.715
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr11_+_112337491
|
0.707
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_+_99391870
|
0.701
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr10_-_60792225
|
0.697
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr18_-_21186107
|
0.695
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr1_-_39871226
|
0.694
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr15_+_78520634
|
0.691
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr15_+_30797466
|
0.684
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr11_+_112337299
|
0.674
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_-_180319013
|
0.674
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr3_-_113842133
|
0.673
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_128257698
|
0.671
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr12_-_119249938
|
0.668
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr2_-_144994038
|
0.662
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr9_+_18464138
|
0.660
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr9_-_35948150
|
0.657
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr6_+_136214495
|
0.657
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr10_-_31328426
|
0.654
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr9_-_74757735
|
0.652
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr3_-_115826481
|
0.651
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr16_+_25135783
|
0.651
|
NM_001169
|
AQP8
|
aquaporin 8
|
|
chr9_-_124280025
|
0.649
|
NM_001004451
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1
|
|
chr4_+_54790020
|
0.646
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr19_+_43616167
|
0.641
|
NM_000540
NM_001042723
|
RYR1
|
ryanodine receptor 1 (skeletal)
|
|
chr4_-_159313167
|
0.641
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_137447479
|
0.637
|
NM_058244
|
WNT8A
|
wingless-type MMTV integration site family, member 8A
|
|
chr2_-_144991386
|
0.636
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_+_6884157
|
0.634
|
NM_001135217
NM_006992
NM_201650
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr6_+_131190237
|
0.629
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr3_-_113842350
|
0.628
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_+_110631916
|
0.626
|
NM_198498
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr1_+_213245507
|
0.625
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr7_+_134226690
|
0.620
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr11_+_113672407
|
0.620
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr12_-_54392328
|
0.609
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr4_-_16509354
|
0.607
|
|
LDB2
|
LIM domain binding 2
|
|
chr1_+_84382562
|
0.607
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_-_90100733
|
0.598
|
NM_001920
|
DCN
|
decorin
|
|
chr3_-_150578103
|
0.596
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr3_-_113842255
|
0.595
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_220788048
|
0.590
|
NM_024746
|
HHIPL2
|
HHIP-like 2
|
|
chr17_+_45598363
|
0.589
|
NM_000023
NM_001135697
|
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein)
|
|
chr7_-_135083906
|
0.584
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr2_+_66519949
|
0.583
|
|
MEIS1
|
Meis homeobox 1
|
|
chr15_+_64782468
|
0.583
|
|
SMAD6
|
SMAD family member 6
|
|
chr20_+_54420574
|
0.580
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr7_+_142629251
|
0.576
|
NM_176882
|
TAS2R40
|
taste receptor, type 2, member 40
|
|
chr4_+_81337663
|
0.572
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr11_+_112337199
|
0.562
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr9_+_117955890
|
0.559
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_-_90100679
|
0.559
|
|
DCN
|
decorin
|
|
chr4_+_41234944
|
0.559
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_44022595
|
0.558
|
|
HOXB3
|
homeobox B3
|
|
chr5_-_39430180
|
0.553
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_90100710
|
0.551
|
|
DCN
|
decorin
|
|
chr2_-_190635566
|
0.546
|
NM_005259
|
MSTN
|
myostatin
|
|
chr8_-_67253234
|
0.546
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr4_-_159311959
|
0.543
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_-_95184221
|
0.534
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr4_+_88973144
|
0.529
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr3_+_35696119
|
0.528
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr5_-_95184121
|
0.527
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr5_+_176449156
|
0.527
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr7_-_75876830
|
0.525
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr8_+_106400322
|
0.524
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr3_-_52454082
|
0.517
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr14_-_36121536
|
0.513
|
NM_014360
|
NKX2-8
|
NK2 homeobox 8
|
|
chr3_+_100840129
|
0.508
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr1_+_84382575
|
0.504
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_+_56888588
|
0.501
|
NM_018488
|
TBX4
|
T-box 4
|
|
chr3_-_113842237
|
0.498
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr17_-_7023378
|
0.496
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr4_+_54790190
|
0.495
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr6_+_88111289
|
0.492
|
NM_001010868
|
C6orf163
|
chromosome 6 open reading frame 163
|
|
chr1_+_162795538
|
0.489
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_-_85103438
|
0.486
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr7_+_143437539
|
0.482
|
NM_001005480
|
OR2A2
|
olfactory receptor, family 2, subfamily A, member 2
|
|
chr13_+_97856148
|
0.480
|
|
|
|
|
chr11_+_48466844
|
0.480
|
NM_001005512
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47
|
|
chr4_+_114433551
|
0.478
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_188935363
|
0.476
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr8_-_124818754
|
0.469
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr12_-_54522881
|
0.466
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr17_+_36493927
|
0.464
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr6_-_119512035
|
0.464
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr8_-_20156971
|
0.463
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr1_+_84402962
|
0.461
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr9_-_72673753
|
0.456
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr5_-_95794416
|
0.453
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr5_-_27074407
|
0.449
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr3_-_38810504
|
0.448
|
NM_006514
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit
|
|
chr1_-_143075560
|
0.448
|
NM_178230
NM_001143883
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4B
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4B
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr9_-_14388981
|
0.447
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_20000938
|
0.443
|
|
NAV2
|
neuron navigator 2
|
|
chr10_+_52504239
|
0.443
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr22_-_16127520
|
0.440
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr13_+_57103789
|
0.439
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr1_-_201321711
|
0.434
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr4_-_24590867
|
0.433
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr7_+_134226727
|
0.432
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_161380562
|
0.432
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr13_+_22653059
|
0.431
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr7_+_100557098
|
0.431
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr1_-_194844061
|
0.430
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr9_-_13240313
|
0.429
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr9_-_21207303
|
0.427
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr1_-_201411533
|
0.427
|
NM_004997
|
MYBPH
|
myosin binding protein H
|
|
chr1_-_149611768
|
0.424
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr17_-_44026321
|
0.424
|
|
HOXB5
|
homeobox B5
|
|
chr2_-_144991556
|
0.424
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144994349
|
0.422
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_-_77801480
|
0.420
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr16_-_88623809
|
0.420
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr7_+_120416103
|
0.419
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr3_+_139211639
|
0.419
|
NM_016369
|
CLDN18
|
claudin 18
|
|
chr7_-_124217978
|
0.419
|
NM_001024603
|
LOC154872
|
hypothetical protein LOC154872
|
|
chr5_-_55564942
|
0.419
|
NM_024669
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chrX_+_10084984
|
0.417
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr11_+_72660894
|
0.417
|
NM_004154
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr4_-_16509126
|
0.416
|
|
LDB2
|
LIM domain binding 2
|
|
chr10_+_50177192
|
0.415
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr5_-_58331515
|
0.415
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_90731671
|
0.413
|
NM_003505
|
FZD1
|
frizzled homolog 1 (Drosophila)
|
|
chr10_-_56230953
|
0.413
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr10_+_11087280
|
0.412
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr21_+_29439768
|
0.412
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_+_108360798
|
0.411
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr4_+_159662349
|
0.410
|
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr17_-_43977273
|
0.408
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr13_+_92677012
|
0.407
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr4_+_154345039
|
0.407
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr5_+_54491702
|
0.405
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr4_-_138673014
|
0.403
|
|
PCDH18
|
protocadherin 18
|
|
chr16_+_31105579
|
0.401
|
|
|
|
|
chr5_-_39460787
|
0.401
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr9_-_83494198
|
0.400
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr3_-_147361626
|
0.399
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr18_+_30651923
|
0.397
|
NM_001198943
|
DTNA
|
dystrobrevin, alpha
|
|
chr20_-_218957
|
0.394
|
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chr2_-_215955206
|
0.393
|
|
FN1
|
fibronectin 1
|
|
chr9_-_14769530
|
0.391
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr6_+_26153590
|
0.391
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr1_+_203503214
|
0.390
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr1_+_146910634
|
0.390
|
NM_001144032
|
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr19_+_614381
|
0.387
|
|
|
|
|
chr4_-_138673078
|
0.386
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr10_-_100985548
|
0.382
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr15_+_66711380
|
0.381
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr2_+_166137091
|
0.380
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr12_-_69434650
|
0.379
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chrX_+_66705407
|
0.378
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr2_+_157822355
|
0.375
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr11_-_55460451
|
0.375
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr21_+_44941514
|
0.373
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr3_+_171621721
|
0.372
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr4_+_120276386
|
0.372
|
NM_016599
|
MYOZ2
|
myozenin 2
|