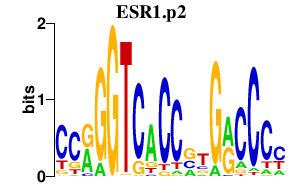
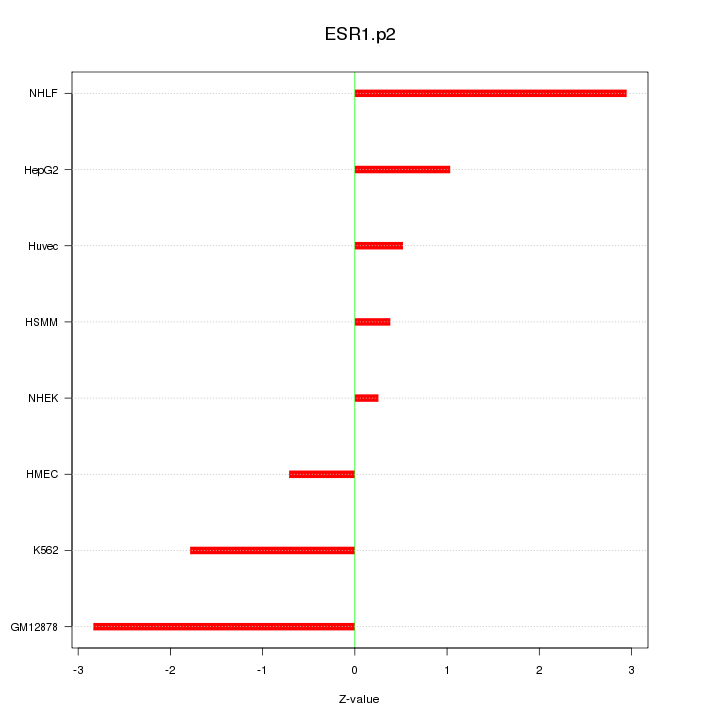
|
chr17_-_24068759
|
2.433
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr19_+_15079141
|
2.255
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr2_+_36436316
|
2.013
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr16_+_54072969
|
1.712
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr22_+_36632242
|
1.682
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr2_-_233501069
|
1.675
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr6_+_148705646
|
1.659
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr17_-_7238546
|
1.630
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr6_+_148705421
|
1.434
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_15955719
|
1.337
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr16_-_1341766
|
1.307
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr9_+_136673464
|
1.303
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr7_+_2638156
|
1.293
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr10_+_115428924
|
1.278
|
NM_033340
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr7_+_99919485
|
1.267
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr9_-_139202855
|
1.264
|
NM_013366
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr19_+_47521600
|
1.219
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr19_-_60802663
|
1.215
|
NM_032836
|
FIZ1
|
FLT3-interacting zinc finger 1
|
|
chr17_-_3518568
|
1.208
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr22_-_18309312
|
1.208
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr2_+_100802923
|
1.199
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr7_-_47588160
|
1.192
|
|
TNS3
|
tensin 3
|
|
chr19_-_15172773
|
1.179
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr16_-_73842884
|
1.177
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr20_+_4077466
|
1.167
|
|
SMOX
|
spermine oxidase
|
|
chr1_+_31659246
|
1.159
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr5_-_156935283
|
1.157
|
|
|
|
|
chr9_-_139202831
|
1.142
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr9_-_83493415
|
1.130
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr2_+_236067424
|
1.112
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr12_-_123023312
|
1.110
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr22_-_36245155
|
1.071
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr19_-_4509471
|
1.070
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr2_-_1727247
|
1.056
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr22_+_42651018
|
1.050
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr16_+_559970
|
1.036
|
NM_004204
NM_148920
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q
|
|
chr12_-_123023052
|
1.035
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr20_+_4077409
|
1.023
|
NM_175839
NM_175840
NM_175841
NM_175842
|
SMOX
|
spermine oxidase
|
|
chr22_-_41446740
|
1.010
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr9_-_85342868
|
1.007
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr9_-_21999270
|
1.004
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr2_+_242398954
|
1.002
|
|
NEU4
|
sialidase 4
|
|
chr1_+_1256585
|
0.998
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr4_+_4912272
|
0.985
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr17_-_3518654
|
0.978
|
NM_014604
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr21_-_45786709
|
0.975
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr19_-_14447163
|
0.967
|
NM_000955
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr1_+_31658549
|
0.963
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr17_-_40401138
|
0.961
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr22_+_42650951
|
0.954
|
NM_025225
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr14_+_92188546
|
0.952
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr22_+_41877502
|
0.946
|
|
TSPO
|
translocator protein (18kDa)
|
|
chr14_+_104244998
|
0.946
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr13_+_112670757
|
0.942
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr17_+_77261401
|
0.937
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr9_+_136673534
|
0.934
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr10_+_102748062
|
0.918
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr22_+_42651007
|
0.917
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr13_+_109757593
|
0.916
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr16_+_88512769
|
0.914
|
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor)
|
|
chr14_+_73073570
|
0.914
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr10_+_50492088
|
0.914
|
NM_001142929
NM_001142933
NM_001142934
NM_020549
|
CHAT
|
choline O-acetyltransferase
|
|
chr2_-_1727220
|
0.913
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr13_-_32822759
|
0.911
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_-_1165106
|
0.907
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr16_-_4105703
|
0.905
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr22_+_41877470
|
0.901
|
NM_000714
NM_007311
|
TSPO
|
translocator protein (18kDa)
|
|
chr3_+_52464657
|
0.898
|
|
NISCH
|
nischarin
|
|
chr12_+_50631734
|
0.894
|
|
ACVR1B
|
activin A receptor, type IB
|
|
chr4_+_151218862
|
0.893
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr2_+_85213884
|
0.889
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr7_+_44110470
|
0.884
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr11_-_1549719
|
0.879
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr19_-_10091534
|
0.854
|
NM_003755
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G
|
|
chr22_+_36472201
|
0.854
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr11_-_70641267
|
0.850
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr20_+_25176698
|
0.842
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr22_-_19122047
|
0.833
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr16_+_14976422
|
0.826
|
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr4_-_118226119
|
0.824
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr19_-_4351357
|
0.818
|
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr15_-_76890626
|
0.816
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr9_-_130980302
|
0.814
|
|
IER5L
|
immediate early response 5-like
|
|
chr17_-_24069299
|
0.813
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr7_+_1536847
|
0.812
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr2_+_106048543
|
0.810
|
NM_032411
|
C2orf40
|
chromosome 2 open reading frame 40
|
|
chr9_-_137731139
|
0.800
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr6_+_148705817
|
0.794
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr16_+_88616508
|
0.790
|
NM_001481
|
GAS8
|
growth arrest-specific 8
|
|
chr5_-_172594691
|
0.786
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr18_+_18003401
|
0.785
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr19_-_11234117
|
0.782
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr9_-_16860664
|
0.776
|
|
BNC2
|
basonuclin 2
|
|
chr5_-_141041933
|
0.771
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr16_+_4361775
|
0.759
|
NM_138440
|
VASN
|
vasorin
|
|
chr22_-_49413326
|
0.759
|
NM_000487
NM_001085425
NM_001085426
NM_001085427
NM_001085428
|
ARSA
|
arylsulfatase A
|
|
chr17_+_77261477
|
0.758
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr17_-_75808680
|
0.756
|
NM_000199
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr19_-_41397396
|
0.752
|
NM_152477
|
ZNF565
|
zinc finger protein 565
|
|
chr9_-_130980359
|
0.746
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr19_-_4486202
|
0.744
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr7_-_47588266
|
0.744
|
|
TNS3
|
tensin 3
|
|
chr19_-_50963942
|
0.742
|
|
|
|
|
chr19_-_5742198
|
0.739
|
NM_001161619
NM_020175
|
DUS3L
|
dihydrouridine synthase 3-like (S. cerevisiae)
|
|
chr17_-_34635446
|
0.739
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr16_+_550394
|
0.737
|
NM_145270
|
C16orf11
|
chromosome 16 open reading frame 11
|
|
chr3_-_129690056
|
0.736
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr6_-_117193505
|
0.729
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr5_-_141041982
|
0.725
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr19_+_42517337
|
0.725
|
NM_181786
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member
|
|
chr17_-_3518628
|
0.721
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr19_+_4255590
|
0.721
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr9_-_138560058
|
0.716
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr2_-_42574587
|
0.715
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr22_-_19122107
|
0.713
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr19_+_3457264
|
0.713
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr22_+_36472176
|
0.712
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr18_+_74841262
|
0.712
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr9_-_139042526
|
0.709
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr17_+_6288457
|
0.707
|
NM_001195228
NM_019013
|
FAM64A
|
family with sequence similarity 64, member A
|
|
chr19_+_7874764
|
0.705
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_+_7834124
|
0.703
|
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr22_-_18309447
|
0.698
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr16_-_68657221
|
0.696
|
|
PDXDC2P
|
pyridoxal-dependent decarboxylase domain containing 2, pseudogene
|
|
chr11_-_605998
|
0.688
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr20_+_61963009
|
0.686
|
NM_080622
|
ABHD16B
|
abhydrolase domain containing 16B
|
|
chr2_-_1727292
|
0.685
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_22909916
|
0.684
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr7_-_100262971
|
0.683
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr11_+_128266460
|
0.679
|
NM_000890
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5
|
|
chr14_-_103251497
|
0.678
|
NM_001100118
NM_001100119
NM_005432
|
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3
|
|
chr19_+_7874765
|
0.675
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr5_+_176446425
|
0.673
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr11_+_832821
|
0.671
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr10_-_133995323
|
0.664
|
|
STK32C
|
serine/threonine kinase 32C
|
|
chr2_+_242398832
|
0.660
|
NM_001167600
NM_001167601
|
NEU4
|
sialidase 4
|
|
chr2_+_236067687
|
0.659
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr17_+_77261440
|
0.659
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr14_+_105012106
|
0.658
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr12_+_50631704
|
0.658
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr16_+_1143738
|
0.654
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr16_+_1974017
|
0.650
|
NM_005262
|
GFER
|
growth factor, augmenter of liver regeneration
|
|
chr1_+_77520190
|
0.647
|
|
AK5
|
adenylate kinase 5
|
|
chr2_-_20714303
|
0.645
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr19_-_41397822
|
0.644
|
NM_001042474
|
ZNF565
|
zinc finger protein 565
|
|
chr19_-_14062230
|
0.642
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr17_+_77261457
|
0.639
|
|
|
|
|
chr7_-_130069399
|
0.638
|
NM_138693
|
KLF14
|
Kruppel-like factor 14
|
|
chr11_-_116213511
|
0.638
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr20_+_30161173
|
0.631
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr17_+_77261447
|
0.630
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr9_-_139042416
|
0.629
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_-_41579325
|
0.629
|
|
|
|
|
chr11_+_47227009
|
0.628
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr1_+_15352800
|
0.628
|
NM_001136218
NM_018022
|
TMEM51
|
transmembrane protein 51
|
|
chr16_-_2125825
|
0.625
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr12_+_50592379
|
0.624
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr7_+_128615948
|
0.620
|
NM_005631
|
SMO
|
smoothened homolog (Drosophila)
|
|
chr10_+_26545241
|
0.616
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr11_+_61204392
|
0.614
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr1_-_151852128
|
0.614
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr11_+_57554977
|
0.613
|
NM_001005186
|
OR6Q1
|
olfactory receptor, family 6, subfamily Q, member 1
|
|
chr20_+_23279372
|
0.610
|
NM_013248
|
NXT1
|
NTF2-like export factor 1
|
|
chr9_+_111850698
|
0.608
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr8_+_144428083
|
0.605
|
|
GLI4
|
GLI family zinc finger 4
|
|
chr1_-_199742871
|
0.605
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr14_+_105063975
|
0.603
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr16_+_1143241
|
0.602
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr22_+_28329544
|
0.602
|
NM_000268
NM_016418
NM_181825
NM_181828
NM_181829
NM_181830
NM_181831
NM_181832
NM_181833
|
NF2
|
neurofibromin 2 (merlin)
|
|
chr1_-_199742864
|
0.602
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr19_-_14447122
|
0.601
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr16_+_64958060
|
0.599
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr19_-_5742177
|
0.597
|
|
DUS3L
|
dihydrouridine synthase 3-like (S. cerevisiae)
|
|
chr1_+_54780515
|
0.594
|
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr19_-_5741913
|
0.591
|
|
DUS3L
|
dihydrouridine synthase 3-like (S. cerevisiae)
|
|
chr9_-_139047112
|
0.591
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chr1_+_179148935
|
0.587
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr19_+_7874762
|
0.585
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr22_+_22445035
|
0.584
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr19_+_62703136
|
0.581
|
|
ZNF773
|
zinc finger protein 773
|
|
chr19_+_58733144
|
0.580
|
NM_001079906
|
ZNF331
|
zinc finger protein 331
|
|
chr19_+_4423149
|
0.579
|
NM_001001520
NM_032631
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr1_-_94475504
|
0.576
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr2_+_219354948
|
0.576
|
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr5_-_156935317
|
0.575
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr1_+_3360880
|
0.575
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr22_-_18309333
|
0.574
|
NM_006440
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr15_-_88034857
|
0.571
|
NM_003847
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha
|
|
chr13_+_112681626
|
0.569
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr20_-_36322587
|
0.569
|
NM_001029864
|
KIAA1755
|
KIAA1755
|
|
chr13_-_109757442
|
0.569
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_-_242096673
|
0.568
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr2_-_216008689
|
0.567
|
|
FN1
|
fibronectin 1
|
|
chr22_-_18309250
|
0.563
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr19_+_3884100
|
0.562
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr1_-_155374756
|
0.558
|
|
ETV3
|
ets variant 3
|
|
chr22_-_35113876
|
0.558
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr2_-_1727324
|
0.557
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr7_+_44110527
|
0.557
|
|
AEBP1
|
AE binding protein 1
|
|
chr11_-_605649
|
0.557
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|