|
chr8_+_110443881
|
2.009
|
NM_177531
|
PKHD1L1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1
|
|
chr1_-_28376041
|
1.633
|
NM_000952
|
PTAFR
|
platelet-activating factor receptor
|
|
chr12_+_20739555
|
1.610
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr12_-_242255
|
1.503
|
NM_001190997
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr2_+_204279442
|
1.493
|
NM_006139
|
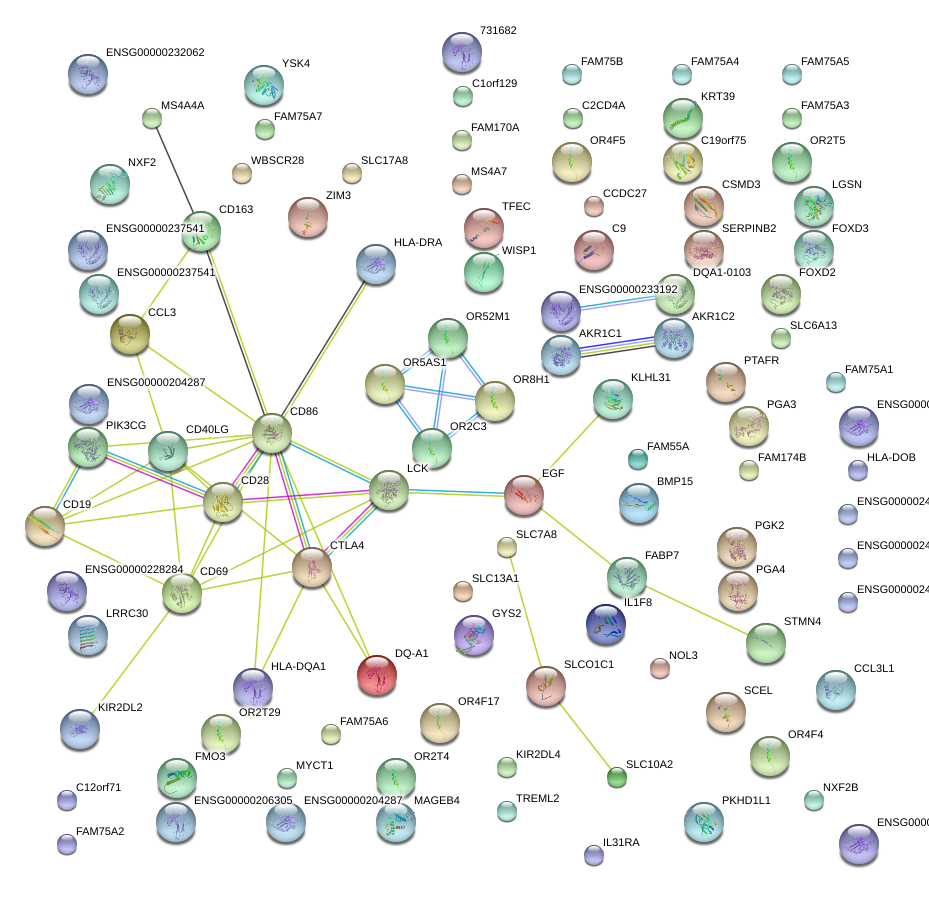
CD28
|
CD28 molecule
|
|
chr6_+_123142559
|
1.414
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr1_+_246591505
|
1.335
|
NM_001004696
|
OR2T4
|
olfactory receptor, family 2, subfamily T, member 4
|
|
chr19_-_62348381
|
1.303
|
NM_052882
|
ZIM3
|
zinc finger, imprinted 3
|
|
chr8_-_114518194
|
1.285
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr16_+_28850760
|
1.278
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr7_-_122627235
|
1.277
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr1_+_169171235
|
1.271
|
NM_001163629
NM_025063
|
C1orf129
|
chromosome 1 open reading frame 129
|
|
chr11_+_59902530
|
1.227
|
NM_021201
NM_206938
NM_206939
NM_206940
|
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7
|
|
chr15_-_91078307
|
1.222
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr8_+_134272463
|
1.201
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr12_-_9804682
|
1.196
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr1_+_3658821
|
1.181
|
NM_152492
|
CCDC27
|
coiled-coil domain containing 27
|
|
chr8_-_27171814
|
1.175
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chrX_-_101581504
|
1.157
|
NM_022053
|
NXF2
|
nuclear RNA export factor 2
|
|
chr2_-_135498716
|
1.156
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chrX_+_135558001
|
1.137
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr6_-_49862858
|
1.122
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr6_+_32713160
|
1.108
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr19_+_56452775
|
1.067
|
NM_173635
|
C19orf75
|
chromosome 19 open reading frame 75
|
|
chrX_+_30169977
|
1.067
|
NM_002367
|
MAGEB4
|
melanoma antigen family B, 4
|
|
chr3_+_123256902
|
1.058
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr10_+_57028755
|
1.043
|
NM_001190478
|
MTRNR2L5
|
MT-RNR2-like 5
|
|
chr11_-_55815113
|
1.035
|
NM_001005199
|
OR8H1
|
olfactory receptor, family 8, subfamily H, member 1
|
|
chr6_-_53638464
|
1.022
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr7_+_72913424
|
1.002
|
NM_182504
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr6_+_153060722
|
0.979
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr11_+_60727559
|
0.963
|
NM_001079807
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr9_+_40690290
|
0.950
|
NM_001083124
|
FAM75A7
FAM75A3
|
family with sequence similarity 75, member A7
family with sequence similarity 75, member A3
|
|
chr18_+_7221136
|
0.943
|
NM_001105581
|
LRRC30
|
leucine rich repeat containing 30
|
|
chr6_-_116529101
|
0.934
|
|
LOC100287673
|
hypothetical LOC100287673
|
|
chr6_-_64087806
|
0.925
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr13_+_77007809
|
0.919
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr12_-_7547552
|
0.910
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr11_-_113935789
|
0.901
|
NM_152315
|
FAM55A
|
family with sequence similarity 55, member A
|
|
chr1_-_245763763
|
0.898
|
NM_198074
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3
|
|
chrX_+_50670474
|
0.887
|
NM_005448
|
BMP15
|
bone morphogenetic protein 15
|
|
chr7_+_106292975
|
0.881
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr1_+_47674275
|
0.880
|
NM_004474
|
FOXD2
|
forkhead box D2
|
|
chr6_+_32515596
|
0.863
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr12_+_99274987
|
0.863
|
NM_001145288
NM_139319
|
SLC17A8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8
|
|
chr19_+_60006877
|
0.854
|
NM_001080770
NM_001080772
NM_002255
|
KIR2DL4
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4
|
|
chr5_+_118993152
|
0.850
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr1_+_32512298
|
0.850
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr5_-_39400319
|
0.848
|
NM_001737
|
C9
|
complement component 9
|
|
chr5_+_55182963
|
0.847
|
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr4_+_111053488
|
0.846
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr1_+_169326641
|
0.841
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr12_-_21649019
|
0.833
|
NM_021957
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr14_-_22722658
|
0.796
|
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr2_-_113526910
|
0.796
|
NM_014438
NM_173178
|
IL1F8
|
interleukin 1 family, member 8 (eta)
|
|
chr7_-_115458064
|
0.789
|
|
TFEC
|
transcription factor EC
|
|
chr1_-_28375722
|
0.783
|
NM_001164723
|
PTAFR
|
platelet-activating factor receptor
|
|
chr17_-_36376663
|
0.782
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr6_-_41276901
|
0.776
|
NM_024807
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chr13_-_102517196
|
0.776
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr12_-_27126721
|
0.774
|
NM_001080406
|
C12orf71
|
chromosome 12 open reading frame 71
|
|
chr11_+_4522993
|
0.773
|
NM_001004137
|
OR52M1
|
olfactory receptor, family 52, subfamily M, member 1
|
|
chr1_+_58899
|
0.773
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr17_-_31441390
|
0.758
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr18_+_59705918
|
0.757
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr11_-_104274606
|
0.754
|
NM_001191016
|
CASP12
|
caspase 12 (gene/pseudogene)
|
|
chr11_+_55554470
|
0.748
|
NM_001001921
|
OR5AS1
|
olfactory receptor, family 5, subfamily AS, member 1
|
|
chr7_+_106293112
|
0.747
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr15_+_60146467
|
0.741
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr1_+_34098662
|
0.729
|
NM_145205
|
HMGB4
|
high-mobility group box 4
|
|
chr6_-_32892705
|
0.728
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr15_+_41672543
|
0.723
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chrX_+_15435351
|
0.720
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr6_+_29732736
|
0.719
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr3_+_98641143
|
0.719
|
NM_173655
|
EPHA6
|
EPH receptor A6
|
|
chr19_+_59820413
|
0.718
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr21_-_31123797
|
0.701
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr8_-_95289945
|
0.701
|
NM_004063
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr8_+_11388918
|
0.699
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr10_-_88707299
|
0.689
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr20_-_46877689
|
0.684
|
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr16_+_49288550
|
0.681
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr10_+_118946989
|
0.681
|
NM_181840
|
KCNK18
|
potassium channel, subfamily K, member 18
|
|
chr9_-_124949033
|
0.676
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_+_152560179
|
0.675
|
NM_080429
|
AQP10
|
aquaporin 10
|
|
chr7_-_38279757
|
0.674
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr1_+_87368044
|
0.670
|
|
LOC339524
|
hypothetical LOC339524
|
|
chr16_-_27806978
|
0.670
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chr7_+_106292932
|
0.669
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr17_-_59437782
|
0.669
|
NM_000873
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr15_-_78050515
|
0.667
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chrX_+_90977115
|
0.660
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr16_+_33168191
|
0.657
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr1_+_86662356
|
0.644
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr20_+_36365965
|
0.642
|
NM_001725
|
BPI
|
bactericidal/permeability-increasing protein
|
|
chr15_-_100280784
|
0.641
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr5_+_156540425
|
0.639
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr16_+_33111656
|
0.638
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr11_-_123772456
|
0.631
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr19_-_16875407
|
0.628
|
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr4_+_41678469
|
0.626
|
NM_001029955
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1
|
|
chr1_+_246617532
|
0.623
|
NM_001005471
|
OR2T6
|
olfactory receptor, family 2, subfamily T, member 6
|
|
chr2_+_73864823
|
0.621
|
NM_001080474
|
C2orf78
|
chromosome 2 open reading frame 78
|
|
chr12_-_83954185
|
0.612
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chrX_+_154880439
|
0.610
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr17_-_35911340
|
0.607
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr2_+_166034402
|
0.603
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr2_-_1905508
|
0.602
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr1_+_151147646
|
0.599
|
NM_005547
|
IVL
|
involucrin
|
|
chr11_+_91724909
|
0.598
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr1_-_156078115
|
0.597
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr3_+_44891101
|
0.595
|
NM_003241
|
TGM4
|
transglutaminase 4 (prostate)
|
|
chr3_-_168853982
|
0.591
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr6_+_160247962
|
0.588
|
NM_002377
|
MAS1
|
MAS1 oncogene
|
|
chr1_-_47468029
|
0.588
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr1_-_32054164
|
0.585
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr5_+_140583098
|
0.581
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr10_-_49792282
|
0.581
|
NM_001006939
|
LRRC18
|
leucine rich repeat containing 18
|
|
chr7_+_133862883
|
0.580
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr4_-_185376012
|
0.580
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr3_+_110024234
|
0.575
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr14_+_75058518
|
0.567
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr12_-_9976173
|
0.562
|
NM_001130711
|
CLEC2A
|
C-type lectin domain family 2, member A
|
|
chr4_-_15549047
|
0.561
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr2_+_102345528
|
0.559
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr12_-_84754289
|
0.558
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr7_+_47661366
|
0.557
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr3_-_46224791
|
0.556
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr17_-_64462976
|
0.542
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr10_-_69125932
|
0.541
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr17_-_53515563
|
0.539
|
|
|
|
|
chr1_-_165754325
|
0.538
|
NM_000734
NM_198053
|
CD247
|
CD247 molecule
|
|
chr8_-_39814860
|
0.533
|
NM_001464
|
ADAM2
|
ADAM metallopeptidase domain 2
|
|
chr1_+_205105776
|
0.530
|
NM_018724
|
IL20
|
interleukin 20
|
|
chr5_+_140568451
|
0.529
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr1_-_150805852
|
0.523
|
NM_178435
|
LCE3E
|
late cornified envelope 3E
|
|
chr6_+_29187565
|
0.515
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr12_+_111829121
|
0.515
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr6_+_37030186
|
0.515
|
NM_153370
|
PI16
|
peptidase inhibitor 16
|
|
chrX_+_101388810
|
0.514
|
NM_022053
|
NXF2
|
nuclear RNA export factor 2
|
|
chr19_-_11425568
|
0.513
|
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr5_-_36036883
|
0.513
|
NM_001171873
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1
|
|
chr10_-_121286034
|
0.512
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr19_-_46080496
|
0.507
|
NM_000764
NM_030589
|
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7
|
|
chr4_-_74704964
|
0.506
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr7_-_149960218
|
0.506
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr22_-_14829803
|
0.503
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chr1_+_246195157
|
0.502
|
NM_001004491
|
OR2AK2
|
olfactory receptor, family 2, subfamily AK, member 2
|
|
chr14_-_75517844
|
0.501
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr15_+_71953020
|
0.497
|
NM_153356
|
TBC1D21
|
TBC1 domain family, member 21
|
|
chr7_+_37746520
|
0.497
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr10_-_17211795
|
0.497
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr2_-_158008763
|
0.496
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr3_-_194579207
|
0.496
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr5_-_83052606
|
0.495
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr12_-_14929991
|
0.495
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr1_+_198263352
|
0.493
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr6_+_168161401
|
0.491
|
NM_005355
NM_030615
|
KIF25
|
kinesin family member 25
|
|
chr19_-_41034734
|
0.489
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr6_-_87861492
|
0.487
|
NM_000735
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr9_-_43570725
|
0.487
|
NM_001145196
|
FAM75A6
|
family with sequence similarity 75, member A6
|
|
chr11_+_48241988
|
0.483
|
NM_001004726
|
OR4X1
|
olfactory receptor, family 4, subfamily X, member 1
|
|
chr6_+_31081707
|
0.478
|
NM_001198815
|
PBMUCL1
|
panbronchiolitis related mucin-like 1
|
|
chrX_+_134693879
|
0.477
|
NM_001017436
NM_152582
|
CT45A3
CT45A4
CT45A2
|
cancer/testis antigen family 45, member A3
cancer/testis antigen family 45, member A4
cancer/testis antigen family 45, member A2
|
|
chr17_+_44642587
|
0.475
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr3_-_46582806
|
0.475
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr7_-_75876830
|
0.472
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr11_-_101292462
|
0.471
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr14_-_75517198
|
0.471
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chrX_-_119095544
|
0.466
|
NM_001099685
NM_032498
|
RHOXF2B
RHOXF2
|
Rhox homeobox family, member 2B
Rhox homeobox family, member 2
|
|
chr6_+_29249289
|
0.463
|
NM_030905
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2
|
|
chr7_+_142053693
|
0.462
|
NM_001190487
|
MTRNR2L6
|
MT-RNR2-like 6
|
|
chr10_+_115929008
|
0.461
|
NM_198795
|
TDRD1
|
tudor domain containing 1
|
|
chr20_+_15125503
|
0.458
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr1_+_207668787
|
0.458
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr2_+_204509698
|
0.457
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr12_+_83954229
|
0.456
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr22_+_36008369
|
0.456
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr7_-_8268666
|
0.454
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr14_+_19780737
|
0.453
|
NM_001004479
|
OR11H4
|
olfactory receptor, family 11, subfamily H, member 4
|
|
chr12_-_84754112
|
0.452
|
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr3_+_85858321
|
0.451
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr12_-_129568362
|
0.451
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr1_-_116112921
|
0.450
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr15_+_84486245
|
0.446
|
NM_152336
|
AGBL1
|
ATP/GTP binding protein-like 1
|
|
chr1_-_104040308
|
0.446
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr5_+_140235114
|
0.445
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr6_-_32447626
|
0.444
|
NM_006781
|
C6orf10
|
chromosome 6 open reading frame 10
|
|
chrX_-_30505804
|
0.442
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr18_+_59405513
|
0.441
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr4_+_130234278
|
0.441
|
NM_173487
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr3_-_194755281
|
0.440
|
NM_032279
|
ATP13A4
|
ATPase type 13A4
|
|
chr13_+_48178620
|
0.440
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chrX_-_131919896
|
0.440
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr21_-_14840506
|
0.438
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr12_-_54150188
|
0.437
|
NM_001005499
|
OR6C70
|
olfactory receptor, family 6, subfamily C, member 70
|
|
chr10_-_48036698
|
0.437
|
NM_016204
|
GDF2
|
growth differentiation factor 2
|
|
chr17_-_25685175
|
0.436
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr2_+_234266250
|
0.436
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr1_+_179269766
|
0.435
|
|
MR1
|
major histocompatibility complex, class I-related
|