|
chr9_-_94226380
|
1.215
|
NM_005014
|
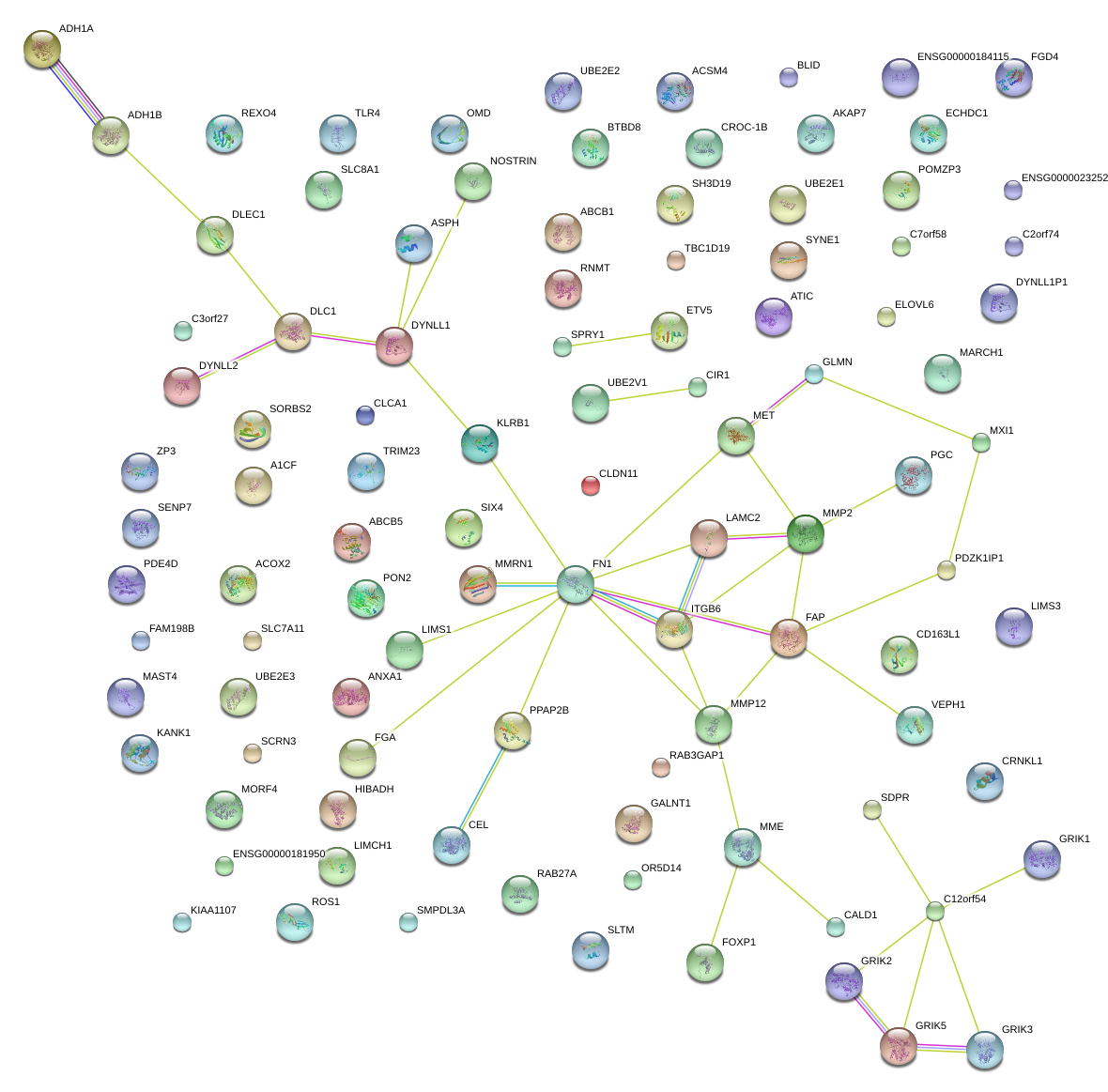
OMD
|
osteomodulin
|
|
chr7_-_94902149
|
0.755
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr1_+_86707113
|
0.691
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr9_+_119506274
|
0.665
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr2_-_192419896
|
0.657
|
|
SDPR
|
serum deprivation response
|
|
chr9_+_119506428
|
0.646
|
|
TLR4
|
toll-like receptor 4
|
|
chr2_-_215951161
|
0.620
|
|
FN1
|
fibronectin 1
|
|
chr9_+_119506446
|
0.618
|
|
TLR4
|
toll-like receptor 4
|
|
chr5_-_59100194
|
0.613
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_58497913
|
0.613
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr10_-_52315390
|
0.607
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr9_+_74956614
|
0.606
|
|
ANXA1
|
annexin A1
|
|
chr4_-_186934059
|
0.584
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_52315436
|
0.573
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr3_+_171621721
|
0.568
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr4_-_100431093
|
0.565
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr12_+_32578513
|
0.562
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr4_-_159311959
|
0.547
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_155731288
|
0.543
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr12_-_7487989
|
0.534
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr7_+_134114957
|
0.530
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_58607673
|
0.524
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_56817823
|
0.521
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_215955206
|
0.519
|
|
FN1
|
fibronectin 1
|
|
chr2_-_192420168
|
0.516
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr7_+_116151411
|
0.509
|
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr3_+_23904086
|
0.508
|
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast)
|
|
chr1_+_92318449
|
0.505
|
NM_183242
|
BTBD8
|
BTB (POZ) domain containing 8
|
|
chr7_+_120415958
|
0.500
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr3_-_71262426
|
0.492
|
|
FOXP1
|
forkhead box P1
|
|
chr4_+_41309690
|
0.481
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_152744143
|
0.480
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr7_+_120416103
|
0.478
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr4_+_26194643
|
0.476
|
NM_018317
|
TBC1D19
|
TBC1 domain family, member 19
|
|
chr1_-_56817717
|
0.473
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_40510923
|
0.469
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr4_-_152368492
|
0.461
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr3_-_187309442
|
0.457
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr16_+_54070302
|
0.441
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_-_160765008
|
0.439
|
|
ITGB6
|
integrin, beta 6
|
|
chr6_-_127705244
|
0.439
|
NM_001139510
|
ECHDC1
|
enoyl CoA hydratase domain containing 1
|
|
chr5_-_64955905
|
0.433
|
NM_001656
NM_033227
NM_033228
|
TRIM23
|
tripartite motif containing 23
|
|
chr1_-_47428310
|
0.433
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr4_+_124540114
|
0.425
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr12_+_7348194
|
0.416
|
NM_001080454
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4
|
|
chr11_-_104274606
|
0.412
|
NM_001191016
|
CASP12
|
caspase 12 (gene/pseudogene)
|
|
chr20_-_19984685
|
0.412
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr4_-_139382672
|
0.407
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chr2_+_215885053
|
0.401
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr9_+_74956601
|
0.396
|
|
ANXA1
|
annexin A1
|
|
chr7_+_134114912
|
0.392
|
|
CALD1
|
caldesmon 1
|
|
chr3_+_156280604
|
0.392
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr11_-_121492010
|
0.390
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr10_+_111975618
|
0.387
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr7_-_87180499
|
0.385
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr11_-_56101772
|
0.384
|
NM_001004741
|
OR5M10
|
olfactory receptor, family 5, subfamily M, member 10
|
|
chr7_+_20621769
|
0.382
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr4_-_111339201
|
0.379
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr14_-_60260211
|
0.376
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr9_+_66293049
|
0.375
|
|
MGC21881
|
hypothetical locus MGC21881
|
|
chr8_-_13178399
|
0.372
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr15_-_53349775
|
0.371
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr18_+_31488530
|
0.370
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr3_-_158703821
|
0.369
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_+_181421796
|
0.369
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr4_-_164754106
|
0.365
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr2_+_61243132
|
0.364
|
NM_001143959
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr6_+_101953581
|
0.362
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr4_-_107508040
|
0.361
|
NM_001195138
|
LOC100507096
|
GTPase IMAP family member-like
|
|
chr6_-_117853710
|
0.359
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr3_-_129777618
|
0.356
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr5_-_58688428
|
0.349
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_75892187
|
0.349
|
NM_001110354
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr8_-_62764839
|
0.343
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr1_+_92405196
|
0.342
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr7_-_27668882
|
0.341
|
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase
|
|
chr3_+_156280539
|
0.335
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_+_135526304
|
0.330
|
NM_001172435
NM_012233
|
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic)
|
|
chr5_+_66160359
|
0.330
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr2_+_169367345
|
0.328
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr2_-_162808123
|
0.327
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr6_+_123152044
|
0.327
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr2_-_160764819
|
0.327
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr10_-_37931864
|
0.326
|
NM_001190489
|
MTRNR2L7
|
MT-RNR2-like 7
|
|
chr2_-_174968595
|
0.321
|
NM_004882
|
CIR1
|
corepressor interacting with RBPJ, 1
|
|
chr3_-_102714663
|
0.318
|
NM_001077203
NM_020654
|
SENP7
|
SUMO1/sentrin specific peptidase 7
|
|
chr2_+_174968701
|
0.311
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr9_+_696744
|
0.310
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_+_74956494
|
0.308
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr4_-_174774368
|
0.306
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr11_+_55319607
|
0.302
|
NM_001004735
|
OR5D14
|
olfactory receptor, family 5, subfamily D, member 14
|
|
chr4_+_91035025
|
0.300
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr2_+_108571336
|
0.300
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr6_-_41823056
|
0.299
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr6_+_131498540
|
0.299
|
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr4_+_87070449
|
0.297
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr4_-_21559471
|
0.296
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chrX_+_102498035
|
0.296
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr2_+_215884923
|
0.296
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr14_-_54720101
|
0.295
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr4_+_86918870
|
0.294
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_+_113436438
|
0.293
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr2_+_190248955
|
0.293
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr1_-_47428190
|
0.291
|
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr5_-_39400319
|
0.290
|
NM_001737
|
C9
|
complement component 9
|
|
chr2_-_20390601
|
0.290
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr5_+_54434230
|
0.287
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr5_-_64955856
|
0.287
|
|
TRIM23
|
tripartite motif containing 23
|
|
chr4_+_146623405
|
0.286
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr2_+_86998248
|
0.286
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr12_+_50342814
|
0.286
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr2_+_108571177
|
0.285
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr6_+_112481970
|
0.284
|
NM_003880
NM_198239
|
WISP3
|
WNT1 inducible signaling pathway protein 3
|
|
chr15_+_53398697
|
0.284
|
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr15_-_40173949
|
0.282
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr4_-_81212737
|
0.280
|
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr11_-_57883117
|
0.279
|
NM_001005489
|
OR5B17
|
olfactory receptor, family 5, subfamily B, member 17
|
|
chr11_+_60765219
|
0.279
|
NM_014224
|
PGA5
|
pepsinogen 5, group I (pepsinogen A)
|
|
chr1_+_25537372
|
0.278
|
NM_014313
|
TMEM50A
|
transmembrane protein 50A
|
|
chr8_+_144802972
|
0.274
|
NM_014789
|
ZNF623
|
zinc finger protein 623
|
|
chr10_+_111957292
|
0.274
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr2_+_215885063
|
0.273
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr3_-_170347019
|
0.272
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr21_-_18697829
|
0.271
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr2_-_112729072
|
0.271
|
NM_032494
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr10_+_104995633
|
0.271
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chr2_+_47483694
|
0.270
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr14_-_68424653
|
0.269
|
|
ACTN1
|
actinin, alpha 1
|
|
chr18_+_6824431
|
0.269
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr6_-_48186868
|
0.269
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr16_-_28528780
|
0.268
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_+_156280772
|
0.267
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_-_153282104
|
0.267
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr6_-_80303822
|
0.266
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr4_-_177353707
|
0.265
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr6_-_122807439
|
0.265
|
|
SERINC1
|
serine incorporator 1
|
|
chr11_-_17147863
|
0.264
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr4_+_41309482
|
0.264
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_+_108571209
|
0.264
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr10_+_70517823
|
0.264
|
NM_002727
|
SRGN
|
serglycin
|
|
chr2_+_138438277
|
0.263
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr2_+_196229715
|
0.263
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr3_-_15814678
|
0.263
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_-_105734535
|
0.262
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr5_-_10360873
|
0.262
|
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr8_-_18585721
|
0.262
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_+_236067424
|
0.262
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chrX_-_153698930
|
0.260
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr2_-_225078931
|
0.260
|
|
CUL3
|
cullin 3
|
|
chr7_+_113513828
|
0.259
|
|
FOXP2
|
forkhead box P2
|
|
chr1_+_149877955
|
0.259
|
|
SNX27
|
sorting nexin family member 27
|
|
chr4_+_86968069
|
0.258
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr12_+_50633430
|
0.258
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr1_+_97033103
|
0.257
|
|
|
|
|
chr3_-_168854395
|
0.257
|
|
WDR49
|
WD repeat domain 49
|
|
chr2_+_207115060
|
0.256
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr6_-_109521969
|
0.255
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr4_+_41309668
|
0.255
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_-_184684478
|
0.255
|
NM_022576
|
PDC
|
phosducin
|
|
chr13_-_35842289
|
0.255
|
NM_001142294
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr11_+_86710076
|
0.255
|
|
TMEM135
|
transmembrane protein 135
|
|
chr2_-_165133239
|
0.254
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr6_+_127940011
|
0.254
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr19_-_14800275
|
0.253
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr6_+_62341966
|
0.253
|
NM_001190706
|
MTRNR2L9
|
MT-RNR2-like 9
|
|
chr3_+_133798770
|
0.253
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr6_-_150099352
|
0.253
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr6_-_152744022
|
0.251
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr11_-_85115105
|
0.251
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_-_106833715
|
0.251
|
NM_152434
|
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe)
|
|
chr4_+_184602778
|
0.251
|
NM_017632
|
CDKN2AIP
|
CDKN2A interacting protein
|
|
chr2_+_174968751
|
0.251
|
|
SCRN3
|
secernin 3
|
|
chr2_-_138762909
|
0.249
|
|
|
|
|
chr1_+_161308318
|
0.248
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr17_-_62793129
|
0.248
|
NM_002816
NM_174871
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr3_-_28365510
|
0.247
|
NM_001134432
NM_001134433
NM_022461
|
AZI2
|
5-azacytidine induced 2
|
|
chr11_+_17289030
|
0.247
|
|
|
|
|
chr1_+_144236398
|
0.247
|
|
ITGA10
|
integrin, alpha 10
|
|
chr12_+_21570457
|
0.245
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr17_-_36719030
|
0.245
|
NM_001146182
|
KRTAP16-1
|
keratin associated protein 16-1
|
|
chr9_-_14076901
|
0.245
|
|
NFIB
|
nuclear factor I/B
|
|
chr18_+_606671
|
0.244
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr6_+_49539049
|
0.243
|
NM_018132
|
CENPQ
|
centromere protein Q
|
|
chr4_+_184602784
|
0.243
|
|
CDKN2AIP
|
CDKN2A interacting protein
|
|
chr5_+_65475760
|
0.242
|
NM_001077199
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr10_+_111959978
|
0.242
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr22_+_31526801
|
0.242
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr19_-_63353956
|
0.242
|
NM_024620
|
ZNF329
|
zinc finger protein 329
|
|
chrX_+_77889861
|
0.241
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr5_-_131160589
|
0.240
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr10_+_123912930
|
0.240
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_+_77209807
|
0.239
|
NM_024684
|
C11orf67
|
chromosome 11 open reading frame 67
|
|
chr10_-_93992982
|
0.239
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr5_+_55098469
|
0.239
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr6_-_31622599
|
0.239
|
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr2_-_215957927
|
0.239
|
|
FN1
|
fibronectin 1
|
|
chr5_+_72287509
|
0.238
|
NM_001146032
NM_138782
|
FCHO2
|
FCH domain only 2
|
|
chr11_+_76491532
|
0.238
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr16_+_2749934
|
0.236
|
|
|
|
|
chr1_+_146910634
|
0.235
|
NM_001144032
|
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|