|
chr2_-_17563186
|
1.950
|
NM_001099218
|
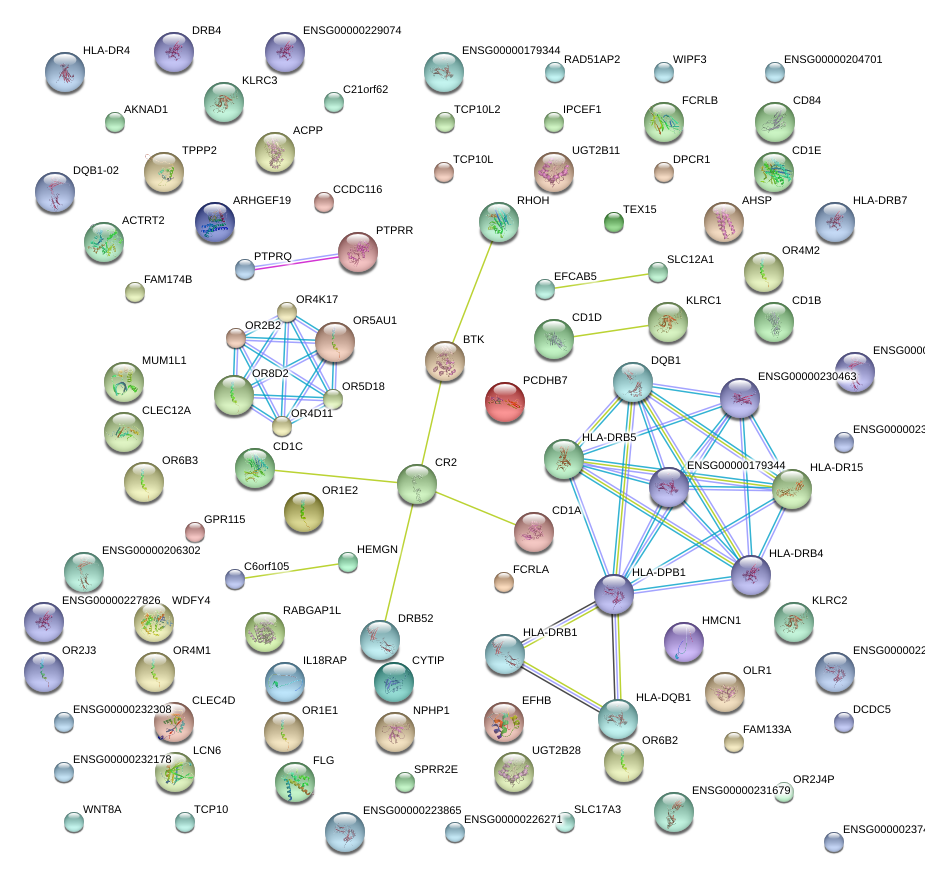
RAD51AP2
|
RAD51 associated protein 2
|
|
chr9_-_70780574
|
1.549
|
|
|
|
|
chrX_-_100527799
|
1.271
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr5_+_137447479
|
0.891
|
NM_058244
|
WNT8A
|
wingless-type MMTV integration site family, member 8A
|
|
chr14_+_19655405
|
0.821
|
NM_001004715
|
OR4K17
|
olfactory receptor, family 4, subfamily K, member 17
|
|
chr6_-_6632197
|
0.771
|
|
|
|
|
chr1_+_183970093
|
0.763
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr12_+_10054492
|
0.757
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chrX_+_92815667
|
0.734
|
NM_001171109
NM_001171110
NM_001171111
NM_173698
|
FAM133A
|
family with sequence similarity 133, member A
|
|
chr12_-_10479810
|
0.720
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr6_-_154719560
|
0.683
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr3_-_19950675
|
0.656
|
NM_144715
|
EFHB
|
EF-hand domain family, member B
|
|
chr11_+_55343658
|
0.617
|
NM_001001952
|
OR5D18
|
olfactory receptor, family 5, subfamily D, member 18
|
|
chr7_+_29840865
|
0.581
|
NM_001080529
|
WIPF3
|
WAS/WASL interacting protein family, member 3
|
|
chr17_-_3283884
|
0.576
|
NM_003554
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2
|
|
chr2_-_158008763
|
0.574
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr21_-_33107813
|
0.565
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_-_109201228
|
0.513
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr4_+_70180805
|
0.511
|
NM_053039
|
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28
|
|
chr3_+_133518900
|
0.509
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr2_+_65517316
|
0.499
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr9_-_99740243
|
0.490
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr9_-_138762725
|
0.484
|
NM_198946
|
LCN6
|
lipocalin 6
|
|
chr10_-_37931864
|
0.482
|
NM_001190489
|
MTRNR2L7
|
MT-RNR2-like 7
|
|
chr1_-_158815895
|
0.481
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr22_+_20316968
|
0.481
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr12_+_8557402
|
0.476
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr15_-_91078307
|
0.469
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr1_+_159959072
|
0.464
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr1_+_156490550
|
0.455
|
NM_001763
|
CD1A
|
CD1a molecule
|
|
chr1_+_205694237
|
0.454
|
NM_001006658
NM_001877
|
CR2
|
complement component (3d/Epstein Barr virus) receptor 2
|
|
chr6_+_47774247
|
0.449
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr15_+_19869841
|
0.437
|
NM_001004719
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2
|
|
chr6_-_25982418
|
0.435
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr12_-_10215942
|
0.431
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr1_-_151333624
|
0.427
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr2_-_110319828
|
0.424
|
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr4_+_39874921
|
0.422
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr11_-_30910074
|
0.408
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr6_-_27988152
|
0.376
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr8_-_30826074
|
0.373
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr11_+_59027624
|
0.370
|
NM_001004706
|
OR4D11
|
olfactory receptor, family 4, subfamily D, member 11
|
|
chr15_+_46285789
|
0.369
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr1_-_93172119
|
0.368
|
|
|
|
|
chr6_-_167717907
|
0.367
|
NM_004610
|
TCP10
|
t-complex 10 homolog (mouse)
|
|
chr10_+_49563520
|
0.367
|
NM_020945
|
WDFY4
|
WDFY family member 4
|
|
chr2_+_102401580
|
0.366
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr6_-_11887265
|
0.364
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chrX_+_105298953
|
0.363
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr2_-_240618518
|
0.359
|
NM_001005853
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2
|
|
chr17_+_25292748
|
0.355
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr5_+_140532385
|
0.345
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr1_+_173111306
|
0.344
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr5_-_138822475
|
0.339
|
NM_001077693
|
ECSCR
|
endothelial cell-specific chemotaxis regulator
|
|
chr14_+_20568184
|
0.336
|
NM_173846
|
TPPP2
|
tubulin polymerization-promoting protein family member 2
|
|
chr1_-_16411690
|
0.327
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr21_-_32879617
|
0.326
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr1_+_2927905
|
0.325
|
NM_080431
|
ACTRT2
|
actin-related protein T2
|
|
chr16_+_31446703
|
0.323
|
NM_016633
|
AHSP
|
alpha hemoglobin stabilizing protein
|
|
chr6_+_31016755
|
0.322
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chr6_-_32665409
|
0.322
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr14_-_20694023
|
0.316
|
NM_001004731
|
OR5AU1
|
olfactory receptor, family 5, subfamily AU, member 1
|
|
chr3_+_133518966
|
0.315
|
|
ACPP
|
acid phosphatase, prostate
|
|
chr12_+_79362256
|
0.313
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr6_+_29187565
|
0.312
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chrX_-_49851743
|
0.307
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr1_-_159099090
|
0.306
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr12_-_10453409
|
0.305
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr6_+_144227265
|
0.304
|
NM_001013623
|
C6orf94
|
chromosome 6 open reading frame 94
|
|
chr11_+_59580705
|
0.303
|
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr1_-_151296611
|
0.298
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr9_-_21067804
|
0.298
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr6_-_32605898
|
0.297
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr3_-_71860958
|
0.297
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr13_+_22653059
|
0.294
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr12_-_7547552
|
0.291
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr17_+_49255237
|
0.287
|
NM_032559
|
KIF2B
|
kinesin family member 2B
|
|
chr7_+_21434818
|
0.284
|
|
|
|
|
chr11_-_112851051
|
0.284
|
NM_000795
NM_016574
|
DRD2
|
dopamine receptor D2
|
|
chr1_+_245779018
|
0.284
|
NM_145278
|
C1orf150
|
chromosome 1 open reading frame 150
|
|
chr8_+_9990471
|
0.283
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr18_+_30427279
|
0.278
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr15_-_78050515
|
0.271
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr6_+_88174408
|
0.269
|
NM_001031743
|
C6orf165
|
chromosome 6 open reading frame 165
|
|
chr3_+_187917791
|
0.266
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr2_-_169596078
|
0.262
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr11_-_49960646
|
0.259
|
NM_001005270
|
OR4C12
|
olfactory receptor, family 4, subfamily C, member 12
|
|
chr1_+_67404756
|
0.254
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr6_+_132915522
|
0.254
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr10_-_91092410
|
0.253
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr17_-_64752550
|
0.252
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr1_-_150070849
|
0.250
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr2_+_108360798
|
0.245
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr20_+_15125503
|
0.242
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_-_21079145
|
0.239
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_-_151310707
|
0.233
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr11_-_55757236
|
0.232
|
NM_001004746
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2
|
|
chr1_+_103961476
|
0.228
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr19_-_14813688
|
0.222
|
NM_001005190
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10
|
|
chr6_+_167504070
|
0.221
|
NM_001145121
|
TCP10L2
TCP10
|
t-complex 10-like 2 (mouse)
t-complex 10 homolog (mouse)
|
|
chr8_+_120148604
|
0.221
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr10_+_96943946
|
0.219
|
NM_207321
|
C10orf129
|
chromosome 10 open reading frame 129
|
|
chr11_+_123315459
|
0.219
|
NM_001001965
|
OR4D5
|
olfactory receptor, family 4, subfamily D, member 5
|
|
chr6_-_29431950
|
0.219
|
NM_030876
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1
|
|
chr12_-_10498481
|
0.216
|
NM_213657
NM_213658
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr17_-_31281398
|
0.216
|
NM_001163122
NM_001163124
NM_001163125
NM_001163130
|
RDM1
|
RAD52 motif 1
|
|
chr5_+_139719970
|
0.215
|
NM_031467
|
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9
|
|
chr11_+_117452936
|
0.214
|
NM_001083947
NM_001173551
NM_001173552
NM_019894
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr7_-_143587653
|
0.208
|
NM_001005328
|
OR2A7
OR2A4
|
olfactory receptor, family 2, subfamily A, member 7
olfactory receptor, family 2, subfamily A, member 4
|
|
chr7_+_94953148
|
0.208
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr13_+_42253685
|
0.207
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr11_-_56914668
|
0.206
|
NM_002728
|
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein)
|
|
chr4_-_70396167
|
0.206
|
NM_021139
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4
|
|
chr5_+_127012611
|
0.198
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr6_+_153060722
|
0.197
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr16_+_56567839
|
0.196
|
NM_199046
NM_199456
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr14_+_21203136
|
0.193
|
NM_001001912
|
OR4E2
|
olfactory receptor, family 4, subfamily E, member 2
|
|
chr7_+_143331954
|
0.193
|
NM_001005281
|
OR6B1
|
olfactory receptor, family 6, subfamily B, member 1
|
|
chr2_+_165860528
|
0.192
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr2_-_240634161
|
0.191
|
NM_173351
|
OR6B3
|
olfactory receptor, family 6, subfamily B, member 3
|
|
chr17_-_25685175
|
0.190
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr10_-_6144119
|
0.189
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr11_+_54867202
|
0.189
|
NM_001005274
|
OR4A16
|
olfactory receptor, family 4, subfamily A, member 16
|
|
chr20_-_7869068
|
0.188
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr3_+_99350859
|
0.188
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr2_-_215605054
|
0.183
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr3_-_166278969
|
0.182
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr11_-_51269023
|
0.180
|
NM_001005272
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5
|
|
chr1_-_115039614
|
0.180
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr18_+_13815542
|
0.180
|
NM_005913
|
MC5R
|
melanocortin 5 receptor
|
|
chr1_+_66570683
|
0.176
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr3_-_64647325
|
0.176
|
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr6_+_54819527
|
0.176
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr1_-_158815850
|
0.175
|
|
CD84
|
CD84 molecule
|
|
chr1_+_190394214
|
0.174
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr11_+_59580630
|
0.174
|
NM_001031666
NM_001031809
NM_006138
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr8_-_139995417
|
0.174
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr17_-_59825221
|
0.173
|
|
|
|
|
chr2_-_110319866
|
0.172
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr21_-_18697829
|
0.171
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr3_-_46761248
|
0.171
|
NM_199183
|
PRSS45
|
protease, serine, 45
|
|
chr2_-_165520241
|
0.170
|
NM_173512
|
SLC38A11
|
solute carrier family 38, member 11
|
|
chr9_-_34371497
|
0.170
|
NM_147168
NM_147169
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr3_+_63613383
|
0.168
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr6_+_28601767
|
0.168
|
NM_001509
NM_003996
|
GPX5
|
glutathione peroxidase 5 (epididymal androgen-related protein)
|
|
chr1_-_150819586
|
0.167
|
NM_032563
|
LCE3D
|
late cornified envelope 3D
|
|
chr7_-_142369534
|
0.165
|
NM_000420
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr1_+_66570363
|
0.163
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr5_-_110090264
|
0.162
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr11_+_55189189
|
0.160
|
NM_001004704
|
OR4C6
|
olfactory receptor, family 4, subfamily C, member 6
|
|
chr5_-_135318420
|
0.160
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr20_-_29360048
|
0.159
|
NM_001037731
|
DEFB116
|
defensin, beta 116
|
|
chr12_-_10173947
|
0.158
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr4_-_145281237
|
0.158
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr4_+_88790477
|
0.157
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr17_-_2913650
|
0.157
|
NM_014566
|
OR1D4
OR1D5
|
olfactory receptor, family 1, subfamily D, member 4 (gene/pseudogene)
olfactory receptor, family 1, subfamily D, member 5
|
|
chr4_-_69116839
|
0.155
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chrX_-_43717864
|
0.155
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr3_-_58538110
|
0.154
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr19_-_44060729
|
0.151
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr11_+_5431213
|
0.150
|
NM_001004754
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2
|
|
chr7_-_142369519
|
0.150
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr11_+_5714253
|
0.149
|
NM_001005180
|
OR56B1
|
olfactory receptor, family 56, subfamily B, member 1
|
|
chr12_-_10433852
|
0.146
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr7_-_121731721
|
0.144
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr11_+_59612712
|
0.144
|
NM_000139
|
MS4A2
|
membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide)
|
|
chr1_-_151614681
|
0.143
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr3_+_28406990
|
0.143
|
NM_001040432
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chrX_-_84250629
|
0.142
|
NM_001012980
|
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1
|
|
chr1_+_201729893
|
0.142
|
NM_014359
|
OPTC
|
opticin
|
|
chr13_+_31211678
|
0.142
|
NM_001166058
NM_130806
|
RXFP2
|
relaxin/insulin-like family peptide receptor 2
|
|
chr3_-_173723754
|
0.141
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr11_+_124329286
|
0.141
|
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr6_-_29503468
|
0.139
|
NM_013937
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1
|
|
chr9_-_103237851
|
0.138
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr17_-_26665220
|
0.135
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_-_168674502
|
0.134
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr1_+_22842704
|
0.131
|
NM_001114101
NM_172369
|
C1QC
|
complement component 1, q subcomponent, C chain
|
|
chr11_+_58967192
|
0.130
|
NM_001004728
|
OR5A1
|
olfactory receptor, family 5, subfamily A, member 1
|
|
chr16_+_2807228
|
0.130
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr8_+_124263932
|
0.130
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr5_-_131907071
|
0.129
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr20_-_31055892
|
0.128
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr9_+_124463665
|
0.128
|
NM_001005236
|
OR1L1
|
olfactory receptor, family 1, subfamily L, member 1
|
|
chr17_+_57801309
|
0.128
|
NM_001144933
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr10_+_124204168
|
0.127
|
NM_001099667
|
ARMS2
|
age-related maculopathy susceptibility 2
|
|
chr6_-_2580296
|
0.127
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr10_-_50012058
|
0.126
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr11_-_5247948
|
0.126
|
NM_005330
|
HBE1
|
hemoglobin, epsilon 1
|
|
chr5_+_156540425
|
0.125
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr11_+_124329222
|
0.123
|
NM_025004
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr2_-_157892391
|
0.122
|
NM_001009959
|
ERMN
|
ermin, ERM-like protein
|
|
chr22_-_31183372
|
0.122
|
NM_174932
|
BPIL2
|
bactericidal/permeability-increasing protein-like 2
|
|
chr12_+_49522967
|
0.122
|
NM_182559
|
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12
|
|
chr17_-_36981830
|
0.121
|
NM_000226
|
KRT9
|
keratin 9
|
|
chr20_-_43693320
|
0.121
|
NM_147198
|
WFDC9
|
WAP four-disulfide core domain 9
|
|
chr11_-_62539857
|
0.120
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr11_-_123758448
|
0.119
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr1_-_68688219
|
0.118
|
NM_000329
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa
|
|
chr6_+_30212488
|
0.118
|
NM_138700
|
TRIM40
|
tripartite motif containing 40
|