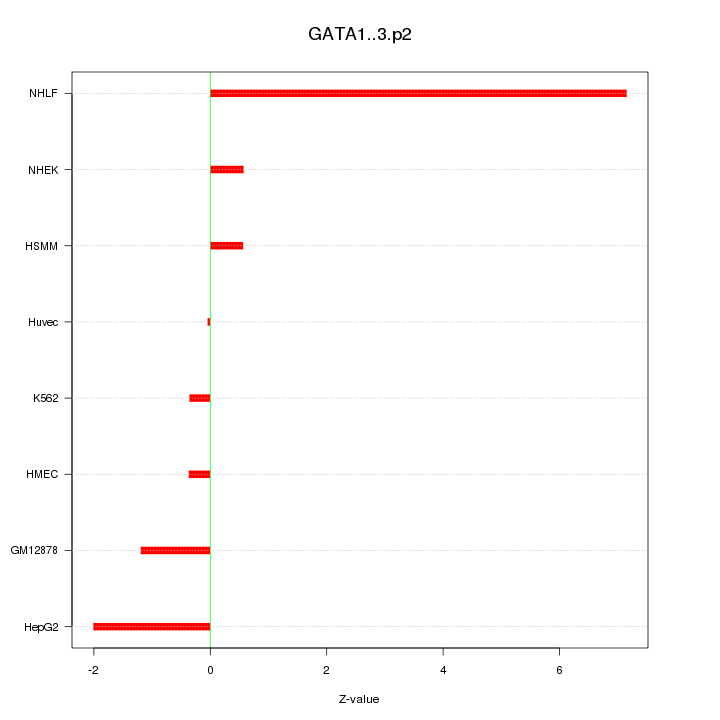
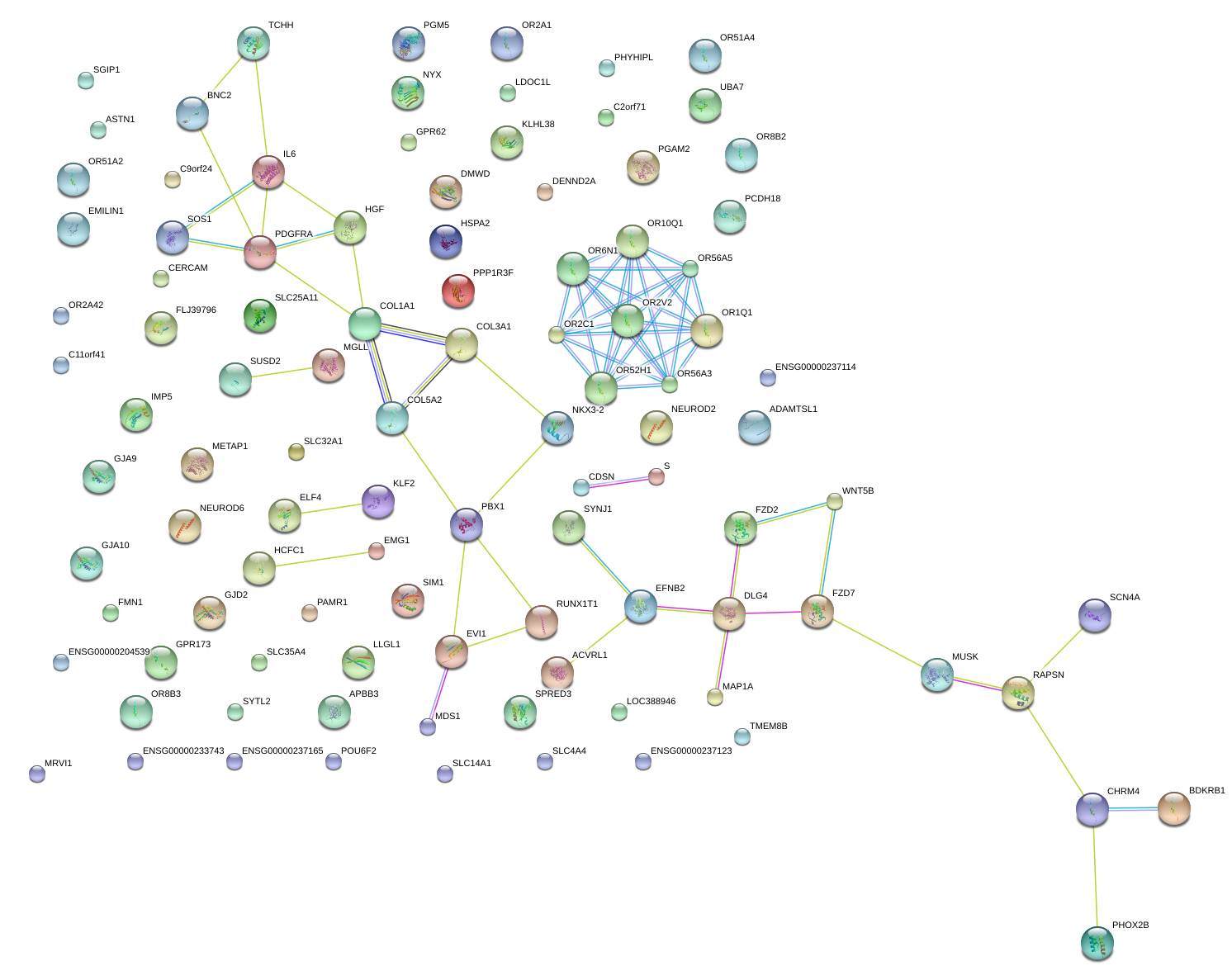
|
chr17_+_39990124
|
6.908
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr1_+_162795538
|
4.493
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_-_35503720
|
4.118
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr12_+_1608672
|
3.595
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr1_+_162795328
|
3.300
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_-_189752575
|
2.999
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr9_-_16717887
|
2.817
|
|
BNC2
|
basonuclin 2
|
|
chr22_+_22907443
|
2.662
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr5_-_71838875
|
2.618
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr9_+_112470871
|
2.540
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr8_-_93176618
|
2.527
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_189547358
|
2.510
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr11_-_5946299
|
2.490
|
NM_001146033
|
OR56A5
|
olfactory receptor, family 56, subfamily A, member 5
|
|
chr15_+_41597097
|
2.404
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr17_+_45617676
|
2.382
|
|
|
|
|
chr9_+_70161634
|
2.380
|
NM_021965
|
PGM5
|
phosphoglucomutase 5
|
|
chr7_-_31347032
|
2.363
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr8_-_93176819
|
2.328
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_152889815
|
2.316
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr3_-_170864111
|
2.276
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr11_-_85146691
|
2.275
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr7_-_81237162
|
2.256
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr7_-_139948780
|
2.069
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr9_+_35819375
|
2.049
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr11_-_47427263
|
2.014
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr2_-_29150630
|
1.991
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr4_-_41445479
|
1.980
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr9_+_35819208
|
1.975
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr22_-_43272100
|
1.959
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr14_+_64077171
|
1.944
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_+_27155197
|
1.912
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chrX_+_53095230
|
1.899
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr3_-_129023850
|
1.899
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr9_+_18464090
|
1.894
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr4_-_41445693
|
1.892
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr17_-_59403932
|
1.881
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr11_+_33520348
|
1.871
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr19_-_50987487
|
1.855
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr14_-_100420936
|
1.850
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chrX_+_49013424
|
1.843
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr2_+_46560207
|
1.835
|
NM_001145051
|
LOC388946
|
transmembrane protein ENSP00000343375
|
|
chr19_+_43572679
|
1.813
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr3_+_51964369
|
1.803
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chr14_+_95792299
|
1.802
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr1_+_66772393
|
1.760
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr9_+_124416768
|
1.752
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr9_-_34387786
|
1.730
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr1_-_157003095
|
1.659
|
NM_001005185
|
OR6N1
|
olfactory receptor, family 6, subfamily N, member 1
|
|
chr15_-_31147376
|
1.657
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr3_-_49826360
|
1.657
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr8_-_93099040
|
1.656
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_189547323
|
1.649
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr16_+_3345889
|
1.645
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chrX_+_41191630
|
1.641
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr4_-_138673078
|
1.639
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_150353179
|
1.631
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr4_+_72423633
|
1.623
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_+_202607554
|
1.612
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr17_-_7047285
|
1.612
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr5_-_139924229
|
1.608
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr20_+_36786518
|
1.599
|
NM_080552
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr17_-_45633873
|
1.597
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr11_-_5523328
|
1.588
|
NM_001005289
|
OR52H1
|
olfactory receptor, family 52, subfamily H, member 1
|
|
chr4_+_54790190
|
1.587
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr1_-_175400665
|
1.576
|
|
ASTN1
|
astrotactin 1
|
|
chr19_+_16296632
|
1.573
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr2_+_189547377
|
1.556
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_-_44071640
|
1.554
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr3_-_49826221
|
1.550
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr6_-_101018271
|
1.537
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr8_-_124734346
|
1.536
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr11_-_4924905
|
1.527
|
NM_001005329
|
OR51A4
|
olfactory receptor, family 51, subfamily A, member 4
|
|
chr4_-_13155135
|
1.514
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr5_+_180514548
|
1.488
|
NM_206880
|
OR2V2
|
olfactory receptor, family 2, subfamily V, member 2
|
|
chr7_-_143560850
|
1.482
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr13_-_105985313
|
1.481
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr11_-_46364681
|
1.455
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr11_+_5925152
|
1.447
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr11_-_10630408
|
1.441
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr9_+_130231027
|
1.428
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr12_+_50592379
|
1.424
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr7_+_38984133
|
1.402
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr11_-_123772456
|
1.384
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr18_+_41560915
|
1.379
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr6_+_90660906
|
1.375
|
NM_032602
|
GJA10
|
gap junction protein, alpha 10, 62kDa
|
|
chr6_-_31196174
|
1.375
|
NM_001264
|
CDSN
|
corneodesmosin
|
|
chr17_-_35017691
|
1.372
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr11_-_57752923
|
1.371
|
NM_001004471
|
OR10Q1
|
olfactory receptor, family 10, subfamily Q, member 1
|
|
chr5_+_139924224
|
1.356
|
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr17_-_4783943
|
1.348
|
NM_001165417
NM_001165418
NM_003562
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11
|
|
chr17_+_41278035
|
1.340
|
NM_175882
|
IMP5
|
intramembrane protease 5
|
|
chr6_-_34209250
|
1.337
|
NM_000841
|
GRM4
|
glutamate receptor, metabotropic 4
|
|
chr1_+_1157485
|
1.333
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chr14_+_68796433
|
1.331
|
NM_001168368
NM_020692
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr18_-_33399601
|
1.330
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr19_+_54669629
|
1.327
|
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr12_+_50342814
|
1.325
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr11_-_64133908
|
1.320
|
|
|
|
|
chr17_-_76064842
|
1.310
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr8_-_93144366
|
1.304
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_27099138
|
1.303
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr9_-_124280025
|
1.300
|
NM_001004451
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1
|
|
chr17_-_43963270
|
1.280
|
NM_002144
|
HOXB1
|
homeobox B1
|
|
chr20_+_36786540
|
1.274
|
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr17_-_24972471
|
1.273
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr22_+_22907417
|
1.260
|
|
SUSD2
|
sushi domain containing 2
|
|
chr17_-_44037332
|
1.259
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chrX_-_110541029
|
1.256
|
NM_000555
|
DCX
|
doublecortin
|
|
chr9_-_35948150
|
1.254
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr4_-_138673014
|
1.253
|
|
PCDH18
|
protocadherin 18
|
|
chr12_+_92595281
|
1.249
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr4_-_123073642
|
1.244
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr12_+_52653176
|
1.235
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr11_-_55460451
|
1.232
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr12_+_79634820
|
1.230
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr22_-_43271843
|
1.224
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr6_-_32268267
|
1.224
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_-_238842011
|
1.214
|
|
GREM2
|
gremlin 2
|
|
chr1_-_200182090
|
1.214
|
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr12_-_51253875
|
1.211
|
NM_175053
|
KRT74
|
keratin 74
|
|
chr8_-_93177057
|
1.210
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_-_105202139
|
1.202
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr5_+_140733664
|
1.202
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr11_-_57171554
|
1.192
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr19_-_54835162
|
1.190
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr8_-_25958291
|
1.187
|
NM_022659
|
EBF2
|
early B-cell factor 2
|
|
chrX_-_57953578
|
1.177
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr17_+_56832492
|
1.170
|
|
TBX2
|
T-box 2
|
|
chr1_-_150079656
|
1.166
|
NM_001136003
|
C2CD4D
|
C2 calcium-dependent domain containing 4D
|
|
chr12_+_50568010
|
1.166
|
NM_001130015
NM_182608
|
ANKRD33
|
ankyrin repeat domain 33
|
|
chr10_+_75343352
|
1.165
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr11_+_48466844
|
1.162
|
NM_001005512
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47
|
|
chr9_-_21207303
|
1.159
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr10_+_70745615
|
1.157
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr10_+_104493701
|
1.153
|
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr17_-_8034071
|
1.152
|
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr1_-_200182338
|
1.152
|
NM_012134
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr19_-_14085979
|
1.152
|
NM_207518
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr11_+_62136788
|
1.152
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr6_-_42527760
|
1.150
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr9_+_18464138
|
1.141
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr5_-_151764857
|
1.137
|
NM_020167
|
NMUR2
|
neuromedin U receptor 2
|
|
chr4_-_87989291
|
1.132
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr4_+_88790477
|
1.130
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr8_-_11096257
|
1.128
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr19_-_14447122
|
1.128
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr19_+_59068787
|
1.125
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr2_-_217248967
|
1.120
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr20_+_1194959
|
1.118
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr11_-_85107569
|
1.116
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_+_170668892
|
1.115
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr5_+_94981735
|
1.115
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr16_-_46738061
|
1.110
|
NM_033226
|
ABCC12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12
|
|
chr19_+_53814359
|
1.109
|
NM_020126
|
SPHK2
|
sphingosine kinase 2
|
|
chr4_+_54790020
|
1.109
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chrX_-_10495476
|
1.100
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr8_-_108579270
|
1.096
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr1_-_116185215
|
1.091
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr17_+_19255083
|
1.089
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr6_-_132952517
|
1.088
|
NM_003967
|
TAAR5
|
trace amine associated receptor 5
|
|
chr9_-_100057809
|
1.078
|
NM_018421
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr19_-_55008172
|
1.076
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr2_-_40510923
|
1.076
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr11_-_57173823
|
1.073
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr19_-_59495949
|
1.072
|
NM_001172654
NM_006865
|
LILRB2
LILRA3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr1_+_147819626
|
1.069
|
NM_178230
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr1_-_238842071
|
1.068
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr3_-_55489973
|
1.067
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr8_+_104900591
|
1.063
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr9_-_34371497
|
1.061
|
NM_147168
NM_147169
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr6_-_80713589
|
1.055
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr5_-_139924013
|
1.052
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr18_-_21185968
|
1.050
|
|
ZNF521
|
zinc finger protein 521
|
|
chr9_-_100057313
|
1.050
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr17_+_56832255
|
1.039
|
|
TBX2
|
T-box 2
|
|
chr10_-_105202012
|
1.039
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr7_-_44195405
|
1.037
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr9_+_139892049
|
1.036
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr11_-_4346191
|
1.036
|
NM_001005161
|
OR52B4
|
olfactory receptor, family 52, subfamily B, member 4
|
|
chr19_+_7506632
|
1.033
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr21_+_46369911
|
1.031
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_-_51114372
|
1.027
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr20_-_47532585
|
1.017
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr22_+_36549334
|
1.016
|
NM_003614
|
GALR3
|
galanin receptor 3
|
|
chrX_-_50403412
|
1.014
|
|
SHROOM4
|
shroom family member 4
|
|
chr19_+_16296734
|
1.013
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr12_-_51086330
|
1.013
|
NM_033033
|
KRT82
|
keratin 82
|
|
chr5_+_140870881
|
1.012
|
|
|
|
|
chr22_+_21011653
|
1.012
|
|
|
|
|
chr8_+_70541631
|
1.008
|
|
SULF1
|
sulfatase 1
|
|
chr15_+_45797977
|
0.997
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr19_+_1389397
|
0.996
|
|
RPS15
|
ribosomal protein S15
|
|
chr19_-_56105805
|
0.990
|
NM_004917
|
KLK4
|
kallikrein-related peptidase 4
|
|
chr12_+_54045161
|
0.989
|
NM_001005497
|
OR6C75
|
olfactory receptor, family 6, subfamily C, member 75
|
|
chr19_-_59383944
|
0.989
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr11_+_91724909
|
0.986
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr13_+_48178620
|
0.985
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr19_+_15713200
|
0.985
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr1_-_26517342
|
0.985
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr5_-_149649380
|
0.984
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|