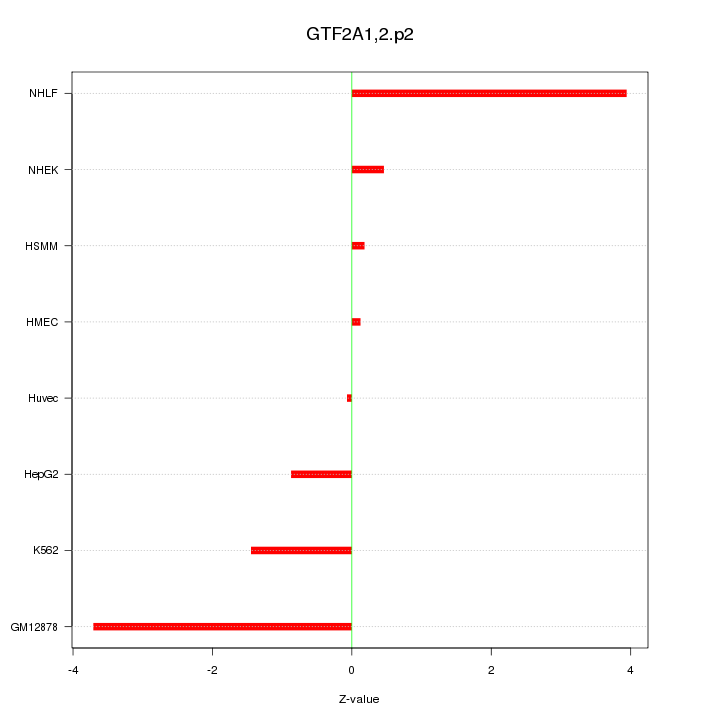
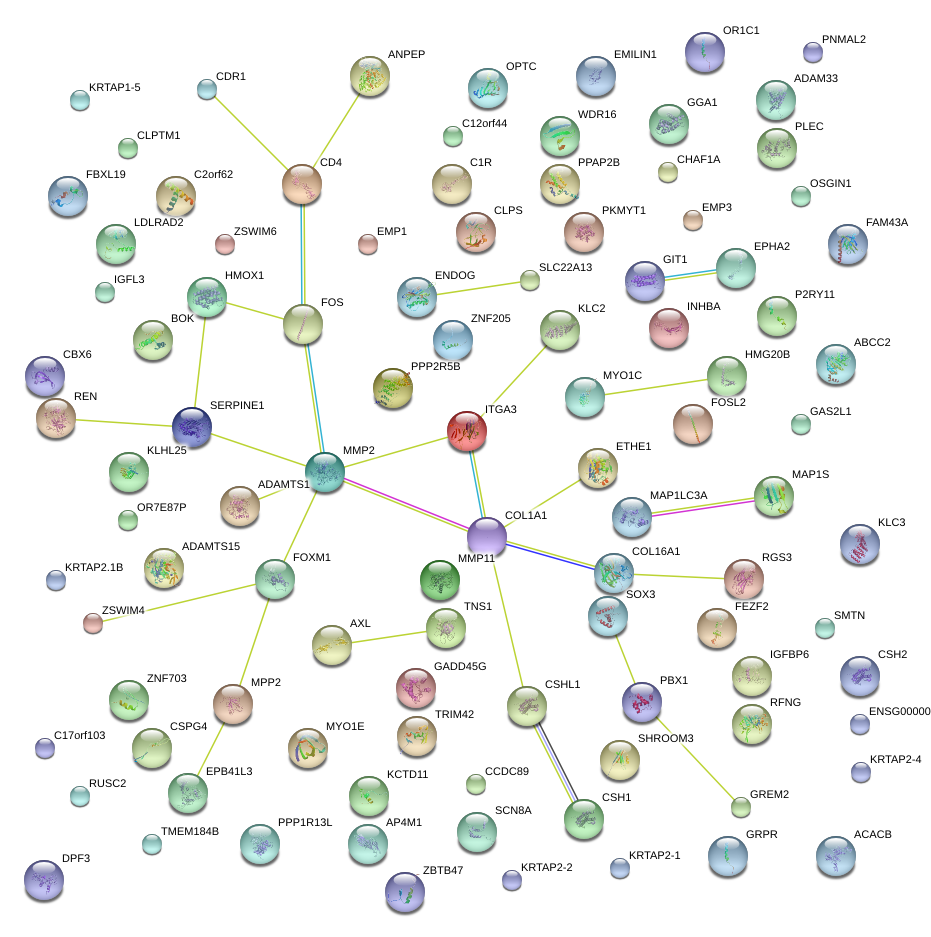
|
chr11_+_129823667
|
4.962
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr7_-_41709191
|
3.787
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr8_+_37672427
|
3.563
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr15_-_73792234
|
3.197
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr15_-_73792150
|
3.082
|
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr2_+_27154938
|
2.862
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr2_+_242146791
|
2.542
|
NM_032515
|
BOK
|
BCL2-related ovarian killer
|
|
chr9_+_35528890
|
2.526
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_-_77602493
|
2.506
|
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr3_+_141879555
|
2.456
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chrX_-_139414890
|
2.387
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr2_+_27155197
|
2.249
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr12_+_13240868
|
2.246
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_-_7136147
|
2.204
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_7136189
|
2.164
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr17_+_45488330
|
2.162
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_-_245988330
|
2.156
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr16_+_54072969
|
2.139
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_+_28469275
|
2.109
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr5_+_60663842
|
2.049
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr17_+_45488356
|
2.010
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr17_-_45633986
|
1.972
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr19_-_50601406
|
1.962
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like
|
|
chr12_+_50342814
|
1.927
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr19_+_46417117
|
1.926
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr14_-_72430545
|
1.908
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr17_-_77602938
|
1.898
|
NM_002917
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chrX_-_139694248
|
1.847
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chr1_-_56817717
|
1.834
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr17_-_36456949
|
1.832
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr7_+_100557098
|
1.831
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr3_-_62334229
|
1.830
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr20_+_32610161
|
1.818
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr2_+_28469850
|
1.815
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr22_-_36998615
|
1.801
|
NM_001195071
|
TMEM184B
|
transmembrane protein 184B
|
|
chr19_-_48723150
|
1.800
|
NM_014297
|
ETHE1
|
ethylmalonic encephalopathy 1
|
|
chr22_-_36998912
|
1.792
|
NM_001195072
NM_012264
|
TMEM184B
|
transmembrane protein 184B
|
|
chr19_+_10083196
|
1.775
|
NM_002566
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11
|
|
chr1_-_56817823
|
1.764
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_3102556
|
1.758
|
NM_001042428
NM_003456
|
ZNF205
|
zinc finger protein 205
|
|
chr19_-_1827133
|
1.749
|
|
|
|
|
chr1_-_202402043
|
1.747
|
NM_000537
|
REN
|
renin
|
|
chr16_+_82544327
|
1.734
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr9_+_35528628
|
1.705
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr22_-_36998889
|
1.692
|
|
TMEM184B
|
transmembrane protein 184B
|
|
chr8_-_145119627
|
1.687
|
NM_201378
|
PLEC
|
plectin
|
|
chr4_+_9094611
|
1.676
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr19_+_46416975
|
1.663
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr10_+_101532450
|
1.654
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr2_+_28469199
|
1.646
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr11_+_64448628
|
1.637
|
NM_006244
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta
|
|
chr19_+_46416971
|
1.628
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr19_-_51690989
|
1.625
|
NM_020709
|
PNMAL2
|
PNMA-like 2
|
|
chr17_-_36436979
|
1.625
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr17_-_39340638
|
1.621
|
NM_005374
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr22_+_29817315
|
1.615
|
|
SMTN
|
smoothelin
|
|
chrX_+_16051344
|
1.612
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr19_+_46417099
|
1.609
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr22_+_28032950
|
1.588
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr17_+_9420668
|
1.564
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chr1_+_22011344
|
1.561
|
NM_001013693
|
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2
|
|
chr1_-_16355013
|
1.556
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr17_-_45633873
|
1.551
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr12_+_108061584
|
1.540
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr9_+_115303527
|
1.523
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_+_99537065
|
1.512
|
NM_004722
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit
|
|
chr2_+_28469864
|
1.508
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr11_-_85074848
|
1.494
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr3_+_42675841
|
1.470
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr12_+_51777715
|
1.444
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr17_-_24940728
|
1.441
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr1_-_31942206
|
1.432
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr1_+_201729893
|
1.432
|
NM_014359
|
OPTC
|
opticin
|
|
chr17_-_1335763
|
1.406
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr17_-_59342319
|
1.404
|
NM_001318
NM_022579
NM_022580
NM_022581
|
CSHL1
|
chorionic somatomammotropin hormone-like 1
|
|
chr22_+_22445035
|
1.397
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr16_-_2970474
|
1.393
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr3_+_38282301
|
1.380
|
NM_004256
|
SLC22A13
|
solute carrier family 22 (organic anion transporter), member 13
|
|
chr12_+_6768934
|
1.368
|
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr1_+_163084707
|
1.364
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr20_-_3610635
|
1.360
|
NM_025220
NM_153202
|
ADAM33
|
ADAM metallopeptidase domain 33
|
|
chr3_+_195887903
|
1.359
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr9_+_130620540
|
1.356
|
NM_004435
|
ENDOG
|
endonuclease G
|
|
chr17_-_21097170
|
1.356
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr19_+_3523903
|
1.348
|
NM_006339
|
HMG20B
|
high-mobility group 20B
|
|
chr19_+_17691302
|
1.347
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr19_+_53520439
|
1.340
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr22_+_36334811
|
1.337
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr1_-_56817568
|
1.331
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_155336223
|
1.331
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr15_-_84139182
|
1.330
|
NM_022480
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr12_+_50750013
|
1.329
|
NM_001098673
NM_021934
|
C12orf44
|
chromosome 12 open reading frame 44
|
|
chr19_+_13767207
|
1.323
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr11_+_65781322
|
1.321
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr19_-_51319769
|
1.320
|
NM_207393
|
IGFL3
|
IGF-like family member 3
|
|
chr15_-_88150823
|
1.314
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr4_+_77829743
|
1.314
|
|
SHROOM3
|
shroom family member 3
|
|
chr21_-_27139563
|
1.311
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr2_+_218929822
|
1.310
|
NM_198559
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chr3_+_50217692
|
1.305
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr15_-_84139021
|
1.294
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr19_+_3523950
|
1.289
|
|
HMG20B
|
high-mobility group 20B
|
|
chr19_+_50150441
|
1.289
|
NM_001294
|
CLPTM1
|
cleft lip and palate associated transmembrane protein 1
|
|
chr16_+_30843396
|
1.285
|
NM_001099784
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr14_+_74815233
|
1.281
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr9_+_91409746
|
1.275
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr6_-_35873034
|
1.274
|
NM_001832
|
CLPS
|
colipase, pancreatic
|
|
chr19_+_50150486
|
1.270
|
|
CLPTM1
|
cleft lip and palate associated transmembrane protein 1
|
|
chr1_-_238842011
|
1.269
|
|
GREM2
|
gremlin 2
|
|
chr19_+_4353658
|
1.268
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr22_+_34107087
|
1.256
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr11_+_65781646
|
1.245
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr22_-_37598115
|
1.244
|
|
CBX6
|
chromobox homolog 6
|
|
chr2_-_218516950
|
1.243
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr1_-_56817802
|
1.243
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr22_-_37598160
|
1.238
|
|
CBX6
|
chromobox homolog 6
|
|
chr8_-_22582526
|
1.236
|
NM_018688
|
BIN3
|
bridging integrator 3
|
|
chr1_-_31942142
|
1.234
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr20_+_59260847
|
1.230
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr12_+_108452117
|
1.226
|
|
UBE3B
|
ubiquitin protein ligase E3B
|
|
chr2_+_176737050
|
1.222
|
NM_006898
|
HOXD3
|
homeobox D3
|
|
chr12_-_123617962
|
1.221
|
NM_001077261
NM_006312
|
NCOR2
|
nuclear receptor corepressor 2
|
|
chr17_-_39340414
|
1.210
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr3_-_50333859
|
1.208
|
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr12_+_6768966
|
1.196
|
|
CD4
|
CD4 molecule
|
|
chr2_+_28469172
|
1.192
|
|
FOSL2
|
FOS-like antigen 2
|
|
chrX_+_134383533
|
1.191
|
|
NCRNA00086
|
non-protein coding RNA 86
|
|
chr22_+_34107051
|
1.181
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr1_+_204710409
|
1.180
|
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr1_+_84540921
|
1.178
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr12_-_51047521
|
1.172
|
NM_002283
|
KRT85
|
keratin 85
|
|
chr11_-_67032125
|
1.166
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr14_+_99859427
|
1.165
|
NM_207117
|
SLC25A47
|
solute carrier family 25, member 47
|
|
chr7_+_72720093
|
1.160
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr19_+_2173415
|
1.153
|
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr1_+_203740391
|
1.152
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr7_-_7541974
|
1.149
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr20_+_62266270
|
1.147
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr16_-_727889
|
1.147
|
|
NARFL
|
nuclear prelamin A recognition factor-like
|
|
chr12_+_51777684
|
1.147
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr17_+_76549658
|
1.144
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr4_-_111763655
|
1.143
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr19_+_17691283
|
1.139
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr16_+_30569558
|
1.138
|
|
PRR14
|
proline rich 14
|
|
chr22_+_36412277
|
1.130
|
NM_024313
|
NOL12
|
nucleolar protein 12
|
|
chr3_-_71262405
|
1.128
|
|
FOXP1
|
forkhead box P1
|
|
chr17_+_74542066
|
1.125
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_-_68470585
|
1.122
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr20_-_43889339
|
1.118
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr6_+_43247168
|
1.113
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr2_+_73973640
|
1.113
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr22_+_31526801
|
1.110
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr21_-_32879617
|
1.108
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr1_+_31655607
|
1.107
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr5_+_150000394
|
1.107
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr15_-_72282452
|
1.102
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr3_+_50279993
|
1.099
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr5_+_150008030
|
1.099
|
|
SYNPO
|
synaptopodin
|
|
chr19_-_5854991
|
1.095
|
NM_001193375
NM_175614
|
NDUFA11
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa
|
|
chr11_-_1549719
|
1.092
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr1_-_151848039
|
1.085
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr1_+_204710453
|
1.080
|
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr1_+_53300410
|
1.080
|
NM_153703
|
PODN
|
podocan
|
|
chr12_+_6768960
|
1.076
|
|
CD4
|
CD4 molecule
|
|
chr17_+_38259620
|
1.076
|
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr1_-_20179495
|
1.075
|
NM_000300
NM_001161727
NM_001161728
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr16_-_48418418
|
1.071
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr1_+_150749943
|
1.063
|
NM_178438
|
LCE5A
|
late cornified envelope 5A
|
|
chr17_-_24940521
|
1.056
|
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr5_+_15553288
|
1.052
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr22_+_20068039
|
1.052
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr7_+_68701704
|
1.044
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr10_+_101532544
|
1.038
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr2_+_94900904
|
1.035
|
NM_144705
|
TEKT4
|
tektin 4
|
|
chr19_+_41085221
|
1.027
|
NM_001007469
NM_014266
|
HCST
|
hematopoietic cell signal transducer
|
|
chr7_+_72720256
|
1.027
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr8_+_42247978
|
1.018
|
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr1_+_203740306
|
1.016
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr1_+_204710208
|
1.014
|
NM_001193321
NM_001193322
NM_014002
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr1_+_149520601
|
1.005
|
|
ZNF687
|
zinc finger protein 687
|
|
chr17_+_7151653
|
1.003
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr19_+_46416938
|
1.000
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr21_+_46367216
|
1.000
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_+_79634820
|
0.999
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr16_-_2970390
|
0.997
|
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr5_+_145296313
|
0.997
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr22_-_40856821
|
0.995
|
NM_000106
NM_001025161
|
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6
|
|
chr22_-_39362569
|
0.995
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr19_-_1430194
|
0.993
|
|
C19orf25
|
chromosome 19 open reading frame 25
|
|
chr17_+_7196151
|
0.988
|
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr10_+_114700775
|
0.988
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr1_+_154351162
|
0.988
|
|
LMNA
|
lamin A/C
|
|
chr16_+_29915003
|
0.987
|
NM_173618
|
INO80E
|
INO80 complex subunit E
|
|
chr19_+_53520632
|
0.980
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr19_-_51081147
|
0.978
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr22_-_29015452
|
0.977
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr22_+_36334400
|
0.973
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr4_+_1330957
|
0.971
|
NM_020894
|
KIAA1530
|
KIAA1530
|
|
chr15_-_87239745
|
0.971
|
NM_178232
|
HAPLN3
|
hyaluronan and proteoglycan link protein 3
|
|
chr22_+_29817218
|
0.970
|
|
SMTN
|
smoothelin
|