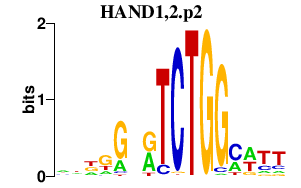
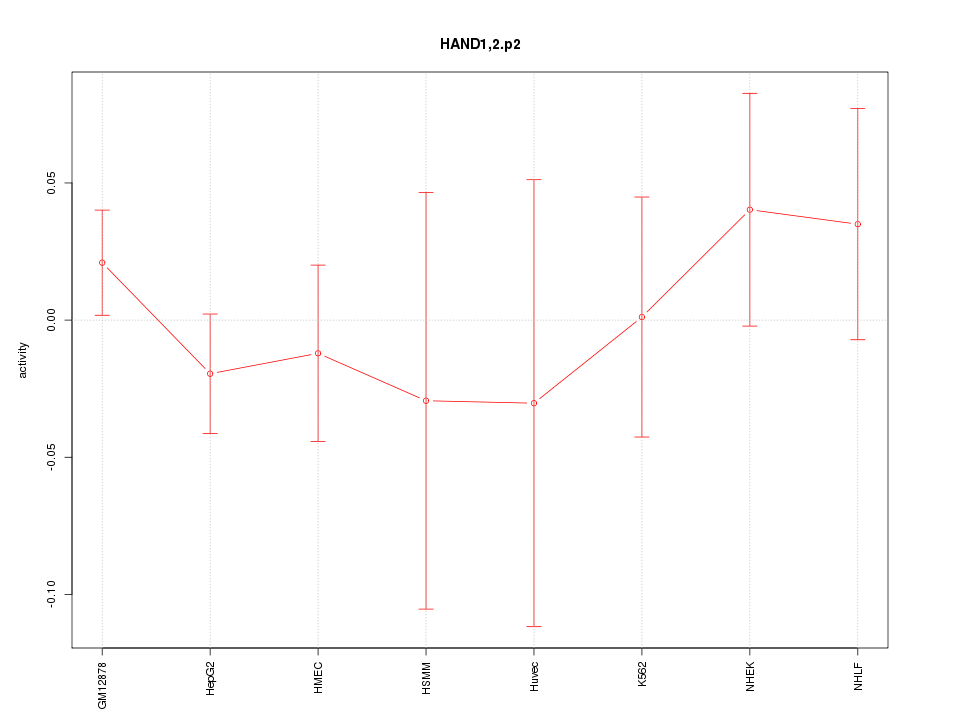
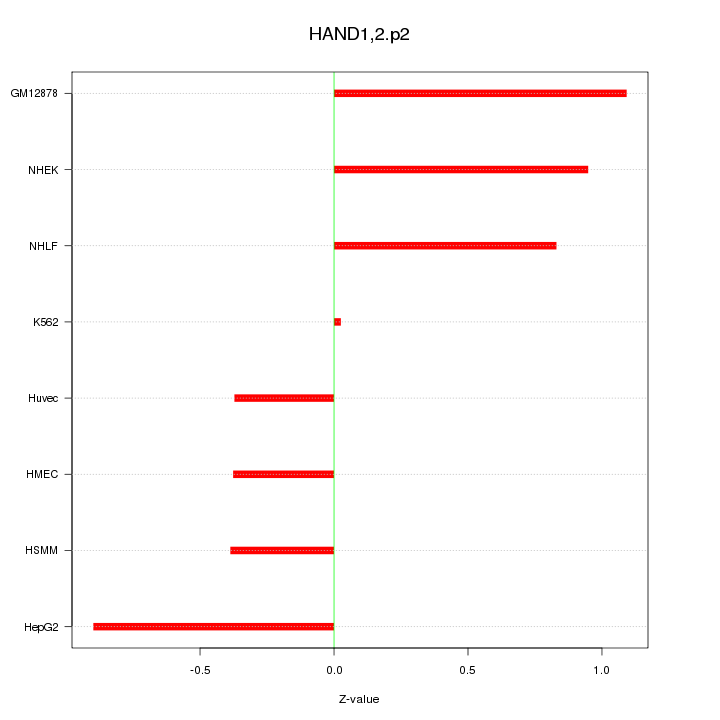
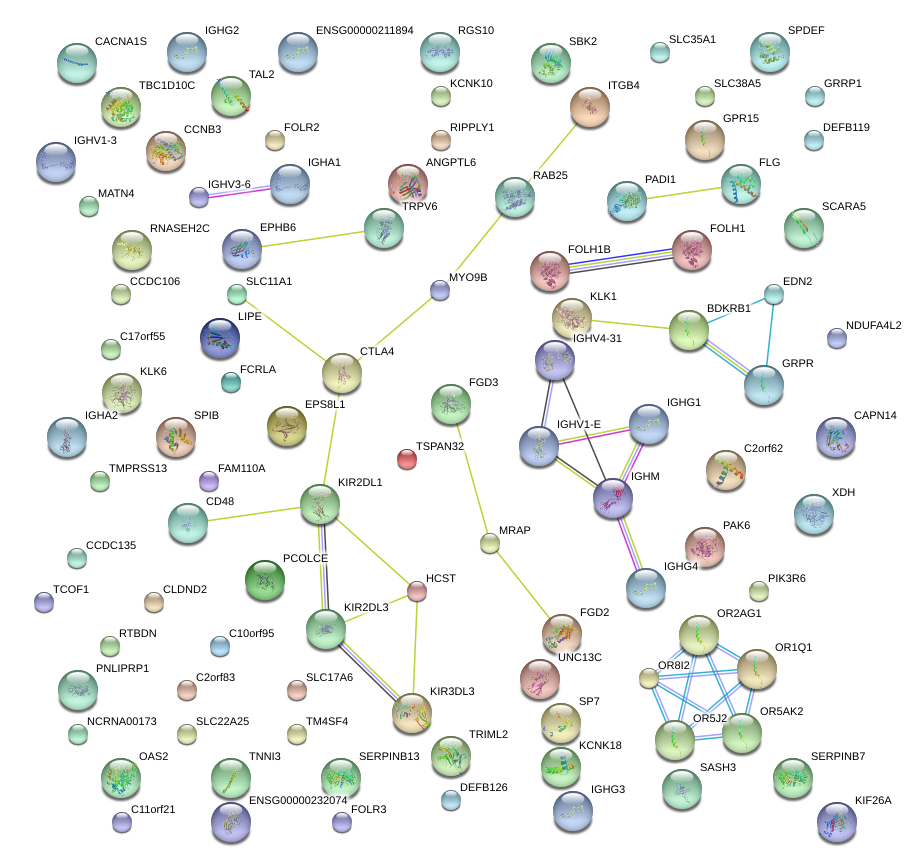
|
chr11_-_62753574
|
2.267
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr11_+_66927976
|
2.206
|
NM_198517
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr2_+_204440753
|
1.734
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr19_+_55614006
|
1.585
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr17_-_8711701
|
1.354
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr18_+_59405513
|
1.347
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr2_-_31293882
|
1.249
|
NM_001145122
|
CAPN14
|
calpain 14
|
|
chr19_+_60279032
|
1.157
|
NM_133180
|
EPS8L1
|
EPS8-like 1
|
|
chr7_+_142270634
|
1.132
|
|
EPHB6
|
EPH receptor B6
|
|
chr17_-_76897590
|
1.129
|
NM_178519
|
C17orf55
|
chromosome 17 open reading frame 55
|
|
chr11_-_65244486
|
1.113
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr20_+_764710
|
1.112
|
NM_207121
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr14_-_87806913
|
1.099
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_+_55617328
|
1.084
|
NM_001003750
|
OR8I2
|
olfactory receptor, family 8, subfamily I, member 2
|
|
chr15_+_52092392
|
1.082
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr11_+_89032112
|
1.017
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr14_-_106241445
|
1.002
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr9_+_124416768
|
0.959
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr12_-_55917134
|
0.944
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr19_-_56163815
|
0.940
|
NM_001012964
NM_001012965
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr11_-_2279642
|
0.931
|
NM_001142946
|
C11orf21
|
chromosome 11 open reading frame 21
|
|
chr19_+_60850765
|
0.930
|
NM_013301
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr6_-_34632061
|
0.929
|
NM_012391
|
SPDEF
|
SAM pointed domain containing ets transcription factor
|
|
chr12_-_50502474
|
0.927
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chr2_+_218929822
|
0.925
|
NM_198559
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chr1_+_180685878
|
0.901
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr6_+_37081387
|
0.895
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr2_-_31491057
|
0.894
|
|
XDH
|
xanthine dehydrogenase
|
|
chr11_+_71605466
|
0.882
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chrX_+_50044279
|
0.874
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr11_+_71524403
|
0.872
|
NM_000804
|
FOLR3
|
folate receptor 3 (gamma)
|
|
chr12_+_115455579
|
0.864
|
|
NCRNA00173
|
non-protein coding RNA 173
|
|
chr1_-_158948176
|
0.863
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chrX_+_128741578
|
0.861
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr10_-_121286034
|
0.861
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chrX_+_128741562
|
0.851
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr11_-_117305277
|
0.842
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr18_+_59571256
|
0.841
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr20_-_29441946
|
0.812
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr1_+_26358097
|
0.805
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr15_+_38318528
|
0.804
|
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr11_+_2279932
|
0.804
|
|
TSPAN32
|
tetraspanin 32
|
|
chr19_-_60360768
|
0.802
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr17_+_71232370
|
0.784
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr16_+_56286212
|
0.783
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr11_+_55700669
|
0.783
|
NM_001005492
|
OR5J2
|
olfactory receptor, family 5, subfamily J, member 2
|
|
chr2_-_228206045
|
0.781
|
NM_001162483
NM_020161
|
C2orf83
|
chromosome 2 open reading frame 83
|
|
chr1_-_41722883
|
0.776
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr10_+_118946989
|
0.758
|
NM_181840
|
KCNK18
|
potassium channel, subfamily K, member 18
|
|
chr1_+_159943385
|
0.756
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr20_-_43370333
|
0.753
|
NM_003833
NM_030590
NM_030592
|
MATN4
|
matrilin 4
|
|
chr11_-_6148131
|
0.751
|
NM_001004052
|
OR52B2
|
olfactory receptor, family 52, subfamily B, member 2
|
|
chr19_-_12807221
|
0.751
|
NM_031429
|
RTBDN
|
retbindin
|
|
chr19_-_47623340
|
0.742
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr1_-_199348316
|
0.735
|
NM_000069
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr19_-_56564068
|
0.730
|
NM_152353
|
CLDND2
|
claudin domain containing 2
|
|
chr1_+_154297588
|
0.726
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr1_+_17404206
|
0.723
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr9_+_107464558
|
0.720
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr2_+_218954995
|
0.714
|
NM_000578
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr11_+_2279812
|
0.707
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr19_+_17047569
|
0.706
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr11_+_22316242
|
0.704
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr12_+_111900650
|
0.702
|
NM_001032731
NM_002535
NM_016817
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa
|
|
chr19_+_59927763
|
0.698
|
NM_153443
|
KIR3DL3
KIR2DL3
KIR3DP1
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 3
killer cell immunoglobulin-like receptor, three domains, pseudogene 1
|
|
chr20_+_71230
|
0.696
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr3_+_99733432
|
0.689
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr7_-_142293382
|
0.686
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr7_+_100037817
|
0.684
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr19_-_10074332
|
0.683
|
NM_031917
|
ANGPTL6
|
angiopoietin-like 6
|
|
chrX_-_48213508
|
0.683
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr10_-_104201234
|
0.681
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr19_-_60740246
|
0.680
|
NM_001101401
|
SBK2
|
SH3-binding domain kinase family, member 2
|
|
chr8_-_27906287
|
0.678
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr11_+_56512922
|
0.676
|
NM_001005323
|
OR5AK2
|
olfactory receptor, family 5, subfamily AK, member 2
|
|
chr12_-_52016270
|
0.673
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr19_+_41085221
|
0.672
|
NM_001007469
NM_014266
|
HCST
|
hematopoietic cell signal transducer
|
|
chrX_+_16051344
|
0.665
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr14_+_95792299
|
0.664
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chrX_-_106033202
|
0.662
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr14_+_103675194
|
0.658
|
|
KIF26A
|
kinesin family member 26A
|
|
chr11_+_6762823
|
0.656
|
NM_001004489
|
OR2AG1
|
olfactory receptor, family 2, subfamily AG, member 1
|
|
chr4_-_189263395
|
0.656
|
NM_173553
|
TRIML2
|
tripartite motif family-like 2
|
|
chr19_-_56585603
|
0.652
|
NM_001193623
|
LOC147646
|
hypothetical protein LOC147646
|
|
chr9_+_94776538
|
0.650
|
NM_033086
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr10_+_118340460
|
0.646
|
NM_006229
|
PNLIPRP1
|
pancreatic lipase-related protein 1
|
|
chr3_+_150675117
|
0.646
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr1_-_158815850
|
0.643
|
|
CD84
|
CD84 molecule
|
|
chr5_+_148766632
|
0.640
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr17_+_3065661
|
0.639
|
NM_014565
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1
|
|
chr11_+_60626505
|
0.639
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr6_-_39390073
|
0.637
|
NM_031460
NM_001135111
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr19_+_61624
|
0.637
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr2_-_31491102
|
0.635
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr14_+_23527866
|
0.634
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr2_+_95621492
|
0.633
|
NM_138800
|
TRIM43
|
tripartite motif containing 43
|
|
chr19_+_59107802
|
0.632
|
NM_031896
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr11_-_133220578
|
0.632
|
NM_174927
|
SPATA19
|
spermatogenesis associated 19
|
|
chr8_-_145809695
|
0.625
|
NM_025251
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr11_+_34599227
|
0.623
|
|
EHF
|
ets homologous factor
|
|
chr17_-_37590910
|
0.618
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr9_-_35818728
|
0.617
|
NM_001012446
|
C9orf128
|
chromosome 9 open reading frame 128
|
|
chr1_-_158815895
|
0.617
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr12_+_53177806
|
0.616
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr19_-_63715135
|
0.612
|
NM_012254
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5
|
|
chr19_-_17819773
|
0.608
|
|
JAK3
|
Janus kinase 3
|
|
chr1_-_29322993
|
0.606
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr1_+_159899528
|
0.605
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr16_-_20246309
|
0.600
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr5_-_149772563
|
0.588
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr12_+_53177789
|
0.583
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr19_-_7702977
|
0.582
|
NM_198492
|
CLEC4G
|
C-type lectin domain family 4, member G
|
|
chrX_+_130505630
|
0.581
|
NM_001004486
|
OR13H1
|
olfactory receptor, family 13, subfamily H, member 1
|
|
chr22_+_23329113
|
0.580
|
NM_001032364
NM_005265
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr19_-_47261761
|
0.577
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr19_+_5865192
|
0.576
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr1_-_207892420
|
0.576
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr8_+_11388918
|
0.573
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr16_+_2450081
|
0.568
|
NM_025108
|
C16orf59
|
chromosome 16 open reading frame 59
|
|
chr11_-_7941617
|
0.567
|
NM_176821
|
NLRP10
|
NLR family, pyrin domain containing 10
|
|
chr19_+_11510578
|
0.565
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr8_+_50987149
|
0.560
|
NM_018967
|
SNTG1
|
syntrophin, gamma 1
|
|
chr4_-_40326603
|
0.559
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_+_246591505
|
0.556
|
NM_001004696
|
OR2T4
|
olfactory receptor, family 2, subfamily T, member 4
|
|
chr11_-_10630408
|
0.550
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr10_-_75080784
|
0.548
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr16_+_639278
|
0.548
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr11_+_62813947
|
0.546
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr17_+_44643067
|
0.546
|
|
ABI3
|
ABI family, member 3
|
|
chr3_+_196933481
|
0.545
|
|
MUC20
|
mucin 20, cell surface associated
|
|
chr3_-_194579207
|
0.541
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr17_-_36409632
|
0.540
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr6_-_25940247
|
0.540
|
NM_005074
|
SLC17A1
|
solute carrier family 17 (sodium phosphate), member 1
|
|
chr22_-_16127520
|
0.540
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr11_+_5558682
|
0.537
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr8_+_1980564
|
0.536
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr20_+_62164900
|
0.533
|
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr6_-_31196174
|
0.533
|
NM_001264
|
CDSN
|
corneodesmosin
|
|
chr16_+_82885821
|
0.531
|
NM_021197
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr9_-_106705867
|
0.531
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr14_-_106240998
|
0.529
|
|
IGHV1-69
IGHG1
|
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr20_+_42776888
|
0.523
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr19_+_60851207
|
0.514
|
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr19_-_56196611
|
0.513
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr19_+_458475
|
0.509
|
NM_033513
|
C19orf20
|
chromosome 19 open reading frame 20
|
|
chr7_+_75977680
|
0.505
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr1_+_26516967
|
0.503
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr16_+_20370359
|
0.499
|
NM_001010845
|
ACSM2A
|
acyl-CoA synthetase medium-chain family member 2A
|
|
chr11_-_62233552
|
0.499
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr22_+_35586950
|
0.499
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr19_+_60691671
|
0.495
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr15_+_61676604
|
0.492
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr1_+_28134090
|
0.491
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr1_+_199519202
|
0.489
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr6_+_146390474
|
0.488
|
NM_000838
NM_001114329
|
GRM1
|
glutamate receptor, metabotropic 1
|
|
chr17_+_20423628
|
0.484
|
NM_001190790
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2
|
|
chr1_+_148511806
|
0.484
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr21_-_42644138
|
0.484
|
NM_005423
|
TFF2
|
trefoil factor 2
|
|
chr1_+_179269766
|
0.480
|
|
MR1
|
major histocompatibility complex, class I-related
|
|
chr22_+_38296703
|
0.478
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chr19_-_56147971
|
0.476
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr14_+_75058518
|
0.473
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr1_+_151035850
|
0.472
|
NM_178352
|
LCE1D
|
late cornified envelope 1D
|
|
chr12_+_39372456
|
0.470
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr5_-_147266257
|
0.469
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr1_-_53972464
|
0.468
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr1_+_34098662
|
0.467
|
NM_145205
|
HMGB4
|
high-mobility group box 4
|
|
chr1_+_200358594
|
0.467
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chrX_+_106030049
|
0.461
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr1_-_150353179
|
0.460
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr17_-_9863929
|
0.457
|
|
GAS7
|
growth arrest-specific 7
|
|
chr17_+_45617676
|
0.456
|
|
|
|
|
chr3_+_51870684
|
0.452
|
NM_203424
|
IQCF2
|
IQ motif containing F2
|
|
chr22_-_29866339
|
0.451
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr2_-_240634161
|
0.447
|
NM_173351
|
OR6B3
|
olfactory receptor, family 6, subfamily B, member 3
|
|
chr3_+_99699137
|
0.445
|
NM_001004737
|
OR5K2
|
olfactory receptor, family 5, subfamily K, member 2
|
|
chr16_-_31054321
|
0.444
|
|
PRSS8
|
protease, serine, 8
|
|
chr8_+_134272463
|
0.441
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr8_-_145661545
|
0.439
|
NM_032687
NM_001129888
|
CYHR1
|
cysteine/histidine-rich 1
|
|
chr5_-_150928697
|
0.438
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chrX_+_36936352
|
0.436
|
NM_001013736
|
FAM47C
|
family with sequence similarity 47, member C
|
|
chr14_-_20694023
|
0.435
|
NM_001004731
|
OR5AU1
|
olfactory receptor, family 5, subfamily AU, member 1
|
|
chr19_-_4351470
|
0.432
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr6_-_55848346
|
0.430
|
|
BMP5
|
bone morphogenetic protein 5
|
|
chr3_-_48576204
|
0.427
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chrX_-_48213545
|
0.427
|
NM_033518
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr12_+_53177758
|
0.426
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr12_+_54045161
|
0.426
|
NM_001005497
|
OR6C75
|
olfactory receptor, family 6, subfamily C, member 75
|
|
chr17_+_38166730
|
0.425
|
NM_005854
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr20_+_16676997
|
0.425
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr6_+_31016755
|
0.423
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chr12_-_10853940
|
0.422
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr14_-_106249999
|
0.422
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr14_-_21172837
|
0.421
|
NM_001005466
|
OR10G2
|
olfactory receptor, family 10, subfamily G, member 2
|
|
chr1_-_110949867
|
0.415
|
NM_004974
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2
|
|
chr12_+_121218218
|
0.415
|
NM_152759
|
LRRC43
|
leucine rich repeat containing 43
|
|
chr3_-_36756355
|
0.404
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chr22_-_38258468
|
0.403
|
|
RPS19BP1
|
ribosomal protein S19 binding protein 1
|
|
chr11_-_111636792
|
0.400
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chr8_+_28230567
|
0.399
|
NM_006228
|
PNOC
|
prepronociceptin
|