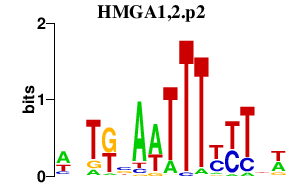
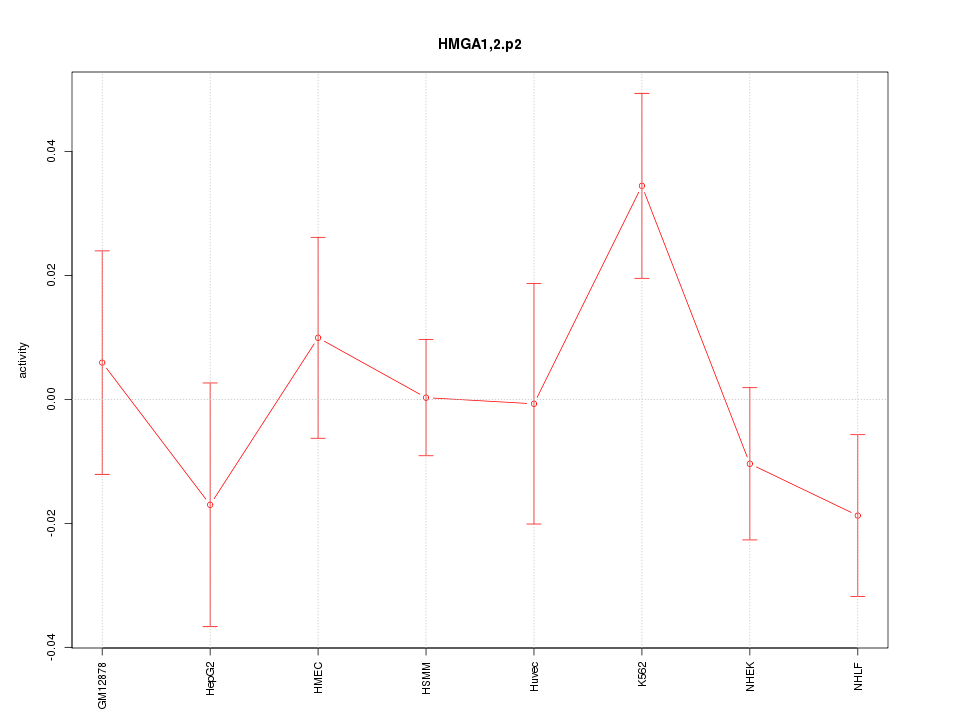
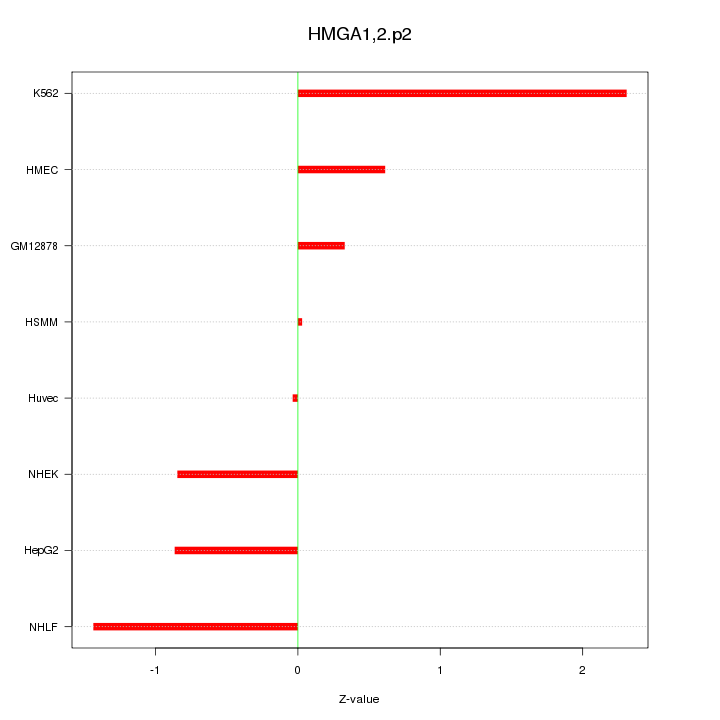
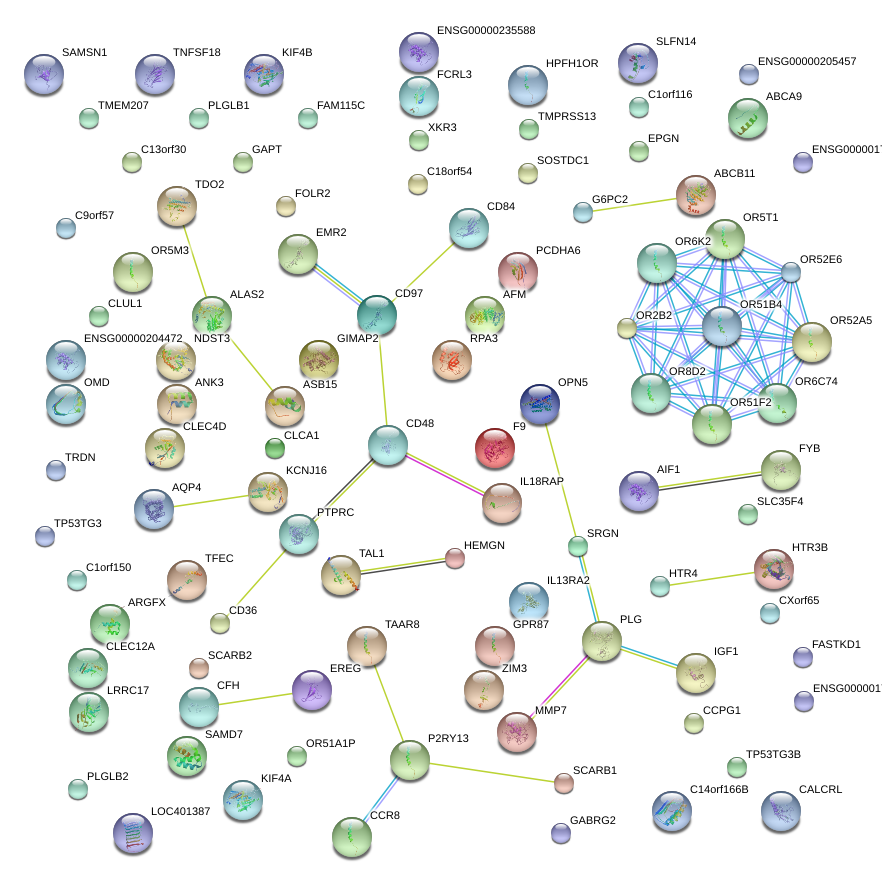
|
chr2_+_169465995
|
3.326
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr7_+_123036312
|
2.605
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr4_+_74566325
|
2.586
|
NM_001133
|
AFM
|
afamin
|
|
chr9_-_94226380
|
2.427
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr12_+_8557402
|
2.224
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr17_-_64568641
|
2.220
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr4_+_75393053
|
2.123
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr6_-_27988152
|
1.925
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr9_+_126460511
|
1.912
|
|
|
|
|
chr2_+_102401580
|
1.818
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr1_+_196874839
|
1.798
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_158815895
|
1.769
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr1_+_196874759
|
1.624
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr19_-_62348381
|
1.508
|
NM_052882
|
ZIM3
|
zinc finger, imprinted 3
|
|
chr18_+_50139168
|
1.500
|
NM_173529
|
C18orf54
|
chromosome 18 open reading frame 54
|
|
chr14_-_57133361
|
1.498
|
NM_001080455
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr7_+_102340579
|
1.456
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr7_-_91632525
|
1.403
|
NM_001161528
|
LOC401387
|
leucine-rich repeat and death domain-containing protein
|
|
chr11_-_101906640
|
1.366
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr1_+_194887630
|
1.365
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr7_-_16471817
|
1.363
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr2_-_87102453
|
1.340
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr7_+_150013716
|
1.329
|
NM_015660
|
GIMAP2
|
GTPase, IMAP family member 2
|
|
chr17_+_65582981
|
1.327
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chrX_-_70243362
|
1.317
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr2_-_169596078
|
1.304
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr9_-_99740243
|
1.286
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr7_-_7724762
|
1.249
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr7_-_112545840
|
1.244
|
|
LOC401397
|
hypothetical LOC401397
|
|
chrX_+_138440560
|
1.243
|
NM_000133
|
F9
|
coagulation factor IX
|
|
chr3_+_122769467
|
1.238
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr12_+_10054492
|
1.227
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chr7_-_112545847
|
1.217
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr2_-_95846109
|
1.208
|
|
LOC150759
|
hypothetical LOC150759
|
|
chr11_-_5130174
|
1.201
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr11_+_4799126
|
1.191
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr11_+_55799604
|
1.153
|
NM_001004745
|
OR5T1
|
olfactory receptor, family 5, subfamily T, member 1
|
|
chr3_+_39346200
|
1.127
|
NM_005201
|
CCR8
|
chemokine (C-C motif) receptor 8
|
|
chrX_-_114158388
|
1.114
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr11_-_123695302
|
1.113
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr1_+_245779018
|
1.102
|
NM_145278
|
C1orf150
|
chromosome 1 open reading frame 150
|
|
chr2_-_188021117
|
1.085
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr12_+_53927248
|
1.078
|
NM_001005490
|
OR6C74
|
olfactory receptor, family 6, subfamily C, member 74
|
|
chr7_-_115458010
|
1.075
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chrX_-_55074125
|
1.066
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr7_+_143043871
|
1.064
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr1_+_196874867
|
1.061
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_+_31690957
|
1.060
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr11_-_117305277
|
1.053
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr10_+_70517863
|
1.045
|
|
SRGN
|
serglycin
|
|
chr1_-_158815850
|
1.043
|
|
CD84
|
CD84 molecule
|
|
chr1_-_158948176
|
1.024
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr11_-_5819702
|
1.015
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr4_+_157044296
|
1.015
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr10_+_70517823
|
1.013
|
NM_002727
|
SRGN
|
serglycin
|
|
chr6_+_47857733
|
1.012
|
NM_181744
|
OPN5
|
opsin 5
|
|
chr10_-_62002672
|
1.008
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_-_14750347
|
1.007
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr5_+_161427225
|
1.005
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr5_+_57823017
|
0.998
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr11_+_113280727
|
0.990
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr3_+_171112053
|
0.979
|
NM_182610
|
SAMD7
|
sterile alpha motif domain containing 7
|
|
chr18_-_22699650
|
0.959
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr21_-_14840506
|
0.954
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr2_-_170138558
|
0.951
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr3_-_152529999
|
0.950
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr5_-_147996769
|
0.946
|
NM_001040169
NM_001040172
NM_001040173
NM_199453
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr22_-_15682555
|
0.931
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr6_+_132915522
|
0.930
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr4_+_119174947
|
0.924
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr5_-_39238818
|
0.915
|
|
FYB
|
FYN binding protein
|
|
chr14_+_76362467
|
0.913
|
NM_194287
|
C14orf166B
|
chromosome 14 open reading frame 166B
|
|
chr11_-_5110383
|
0.909
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chr1_-_205272619
|
0.905
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr11_-_5279750
|
0.893
|
NM_033179
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4
|
|
chr5_+_140227951
|
0.889
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr1_-_156937065
|
0.882
|
NM_001005279
|
OR6K2
|
olfactory receptor, family 6, subfamily K, member 2
|
|
chr9_-_73865318
|
0.876
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr17_+_65612589
|
0.872
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr12_-_101396459
|
0.871
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_155936904
|
0.869
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr1_-_171286634
|
0.864
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr15_-_53440069
|
0.856
|
|
CCPG1
|
cell cycle progression 1
|
|
chr11_+_71605466
|
0.856
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr7_-_115458064
|
0.855
|
|
TFEC
|
transcription factor EC
|
|
chr3_-_152517204
|
0.853
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr1_+_61315378
|
0.852
|
|
|
|
|
chr18_+_606671
|
0.851
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr1_+_86707113
|
0.848
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr5_+_154373452
|
0.846
|
NM_001099293
|
KIF4B
|
kinesin family member 4B
|
|
chr6_-_123999640
|
0.839
|
NM_006073
|
TRDN
|
triadin
|
|
chr17_-_30909219
|
0.834
|
NM_001129820
|
SLFN14
|
schlafen family member 14
|
|
chr11_-_55994548
|
0.831
|
NM_001004742
|
OR5M3
|
olfactory receptor, family 5, subfamily M, member 3
|
|
chr16_+_33111656
|
0.830
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr13_+_42253685
|
0.829
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr4_+_75449720
|
0.822
|
NM_001432
|
EREG
|
epiregulin
|
|
chr3_-_191650291
|
0.820
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr7_+_80105718
|
0.816
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_-_47468029
|
0.815
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr1_-_160648429
|
0.811
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr12_-_9976173
|
0.805
|
NM_001130711
|
CLEC2A
|
C-type lectin domain family 2, member A
|
|
chr6_+_28032997
|
0.805
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr5_-_20024050
|
0.803
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr5_-_90645974
|
0.800
|
|
LOC100505994
|
hypothetical LOC100505994
|
|
chr5_-_88155360
|
0.795
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_-_87311624
|
0.794
|
NM_138817
|
SLC7A13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13
|
|
chr17_+_25292748
|
0.793
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr1_+_185064654
|
0.788
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr7_+_141054621
|
0.785
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr11_+_59804634
|
0.785
|
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chrX_+_115215985
|
0.783
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr12_-_83954185
|
0.778
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr1_-_167969802
|
0.777
|
NM_000450
|
SELE
|
selectin E
|
|
chr13_-_45577144
|
0.777
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr17_-_64752550
|
0.775
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr5_+_40945315
|
0.773
|
NM_000587
|
C7
|
complement component 7
|
|
chr7_-_126670804
|
0.772
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr7_-_113346284
|
0.771
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr10_-_17211795
|
0.771
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr1_-_184696839
|
0.770
|
NM_002597
|
PDC
|
phosducin
|
|
chr4_-_74704964
|
0.769
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr12_+_18305740
|
0.769
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr10_+_74323344
|
0.769
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr4_-_38710376
|
0.766
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr6_+_14032655
|
0.760
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr10_+_115514567
|
0.757
|
NM_182601
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr17_-_6924227
|
0.753
|
NM_006344
NM_182906
|
CLEC10A
|
C-type lectin domain family 10, member A
|
|
chr6_-_53638464
|
0.748
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr6_-_68655708
|
0.746
|
|
|
|
|
chr4_-_70396167
|
0.742
|
NM_021139
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4
|
|
chr1_-_245902965
|
0.741
|
NM_001005487
|
OR13G1
|
olfactory receptor, family 13, subfamily G, member 1
|
|
chr19_-_9098625
|
0.736
|
NM_001001958
|
OR7G3
|
olfactory receptor, family 7, subfamily G, member 3
|
|
chr5_-_121842680
|
0.733
|
|
|
|
|
chr3_+_157321030
|
0.732
|
NM_172160
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr3_-_36756355
|
0.732
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chrX_-_62487693
|
0.731
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr3_+_115099007
|
0.730
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr5_+_94752803
|
0.725
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr14_+_20319049
|
0.723
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr1_-_143786993
|
0.722
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_-_68379331
|
0.722
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr5_+_40945374
|
0.720
|
|
C7
|
complement component 7
|
|
chr11_+_26310253
|
0.719
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr5_+_170143289
|
0.718
|
NM_014211
|
GABRP
|
gamma-aminobutyric acid (GABA) A receptor, pi
|
|
chr2_+_157822355
|
0.706
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr5_-_131907071
|
0.704
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr7_+_123353144
|
0.704
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr18_-_22696532
|
0.704
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chrX_+_134693879
|
0.699
|
NM_001017436
NM_152582
|
CT45A3
CT45A4
CT45A2
|
cancer/testis antigen family 45, member A3
cancer/testis antigen family 45, member A4
cancer/testis antigen family 45, member A2
|
|
chr15_+_56217686
|
0.697
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr1_+_78858675
|
0.697
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr10_+_97749837
|
0.696
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr1_-_76170563
|
0.693
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chrX_-_12972641
|
0.688
|
NM_174901
|
FAM9C
|
family with sequence similarity 9, member C
|
|
chr7_-_135263737
|
0.687
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr12_-_9804682
|
0.683
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr1_+_171871283
|
0.681
|
NM_001195190
|
LOC730159
|
hypothetical protein LOC730159
|
|
chr3_-_167037946
|
0.680
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr11_+_63032034
|
0.680
|
NM_001142537
NM_001142538
|
LGALS12
|
lectin, galactoside-binding, soluble, 12
|
|
chr5_-_39400319
|
0.680
|
NM_001737
|
C9
|
complement component 9
|
|
chr1_+_111571803
|
0.676
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr16_+_33168191
|
0.676
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr7_+_50106227
|
0.670
|
NM_001161834
|
C7orf72
|
chromosome 7 open reading frame 72
|
|
chr12_-_94469357
|
0.669
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr6_+_34590624
|
0.669
|
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr11_-_102001259
|
0.667
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr12_+_21570457
|
0.666
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr6_-_87861492
|
0.665
|
NM_000735
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr14_+_59934178
|
0.662
|
|
|
|
|
chr12_+_10015274
|
0.660
|
NM_138337
NM_201623
|
CLEC12A
|
C-type lectin domain family 12, member A
|
|
chr12_-_49076658
|
0.660
|
NM_001145475
|
FAM186A
|
family with sequence similarity 186, member A
|
|
chr11_-_32772731
|
0.656
|
NM_001008391
|
CCDC73
|
coiled-coil domain containing 73
|
|
chr6_+_154449334
|
0.655
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr10_+_118177371
|
0.654
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr2_-_178495768
|
0.654
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr20_-_30703219
|
0.653
|
NM_182584
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr5_-_83052606
|
0.653
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr12_-_53975282
|
0.652
|
NM_001005493
|
OR6C6
|
olfactory receptor, family 6, subfamily C, member 6
|
|
chr17_-_72151314
|
0.651
|
NM_018414
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
|
|
chr11_-_107969554
|
0.650
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr4_-_88669625
|
0.650
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr19_-_7009644
|
0.646
|
NM_001164425
|
MBD3L3
|
methyl-CpG-binding domain protein 3-like 3-like
|
|
chr6_-_32447626
|
0.645
|
NM_006781
|
C6orf10
|
chromosome 6 open reading frame 10
|
|
chr5_+_140481763
|
0.644
|
NM_018938
|
PCDHB4
|
protocadherin beta 4
|
|
chr1_-_191422346
|
0.642
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr21_-_18697829
|
0.642
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr2_-_136590210
|
0.639
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr7_+_121300394
|
0.638
|
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr1_+_195503956
|
0.636
|
NM_001193640
NM_201253
|
CRB1
|
crumbs homolog 1 (Drosophila)
|
|
chr18_-_59480082
|
0.634
|
NM_006919
|
SERPINB3
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr19_+_14554895
|
0.633
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chrX_+_105298953
|
0.632
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr1_+_65492679
|
0.631
|
|
LOC645195
DNAJC6
|
hypothetical LOC645195
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr8_-_82769761
|
0.627
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr19_+_60168463
|
0.623
|
NM_001174081
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chrX_+_79562601
|
0.622
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr4_-_139382672
|
0.621
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chrX_-_80344096
|
0.618
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr7_+_21434818
|
0.616
|
|
|
|
|
chrX_+_21868635
|
0.613
|
NM_004595
|
SMS
|
spermine synthase
|