|
chr14_+_62740878
|
1.496
|
NM_020663
|
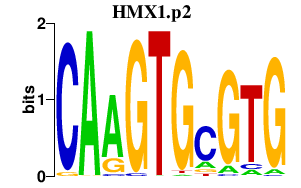
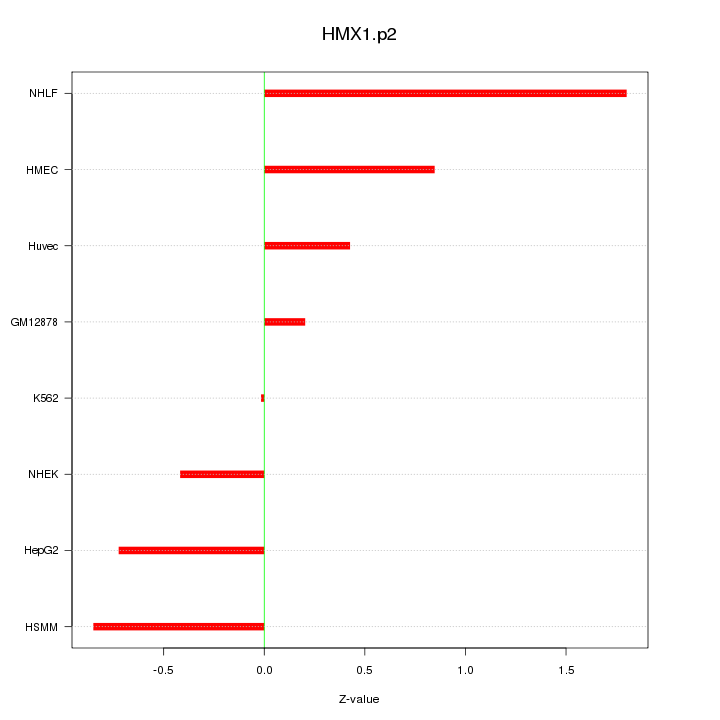
RHOJ
|
ras homolog gene family, member J
|
|
chr20_-_41251878
|
1.255
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr16_-_85099941
|
1.093
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr8_-_26780665
|
1.088
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr11_+_69602055
|
1.078
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr18_-_44189625
|
1.056
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr17_+_19255083
|
0.917
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr5_-_168660293
|
0.879
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr7_+_128257698
|
0.867
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr21_-_38954433
|
0.809
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr2_+_100087812
|
0.799
|
|
|
|
|
chr1_+_155130146
|
0.750
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr20_-_21442663
|
0.741
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr7_+_5609416
|
0.739
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr2_-_241408296
|
0.718
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr1_-_175400441
|
0.715
|
|
ASTN1
|
astrotactin 1
|
|
chr18_-_55091596
|
0.711
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr1_-_27159462
|
0.702
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chrX_-_142550535
|
0.663
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr5_-_10406690
|
0.656
|
|
|
|
|
chr1_-_32002222
|
0.651
|
NM_001703
|
BAI2
|
brain-specific angiogenesis inhibitor 2
|
|
chr8_+_120497732
|
0.649
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr1_+_12149637
|
0.648
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr4_+_2390469
|
0.647
|
NM_001193282
|
LOC402160
|
hypothetical protein LOC402160
|
|
chr13_-_113651815
|
0.643
|
NM_182614
|
FAM70B
|
family with sequence similarity 70, member B
|
|
chr9_+_134843918
|
0.624
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chrX_+_37429927
|
0.614
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr14_+_100370559
|
0.608
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr19_-_14467890
|
0.601
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr16_-_716367
|
0.590
|
NM_001031737
|
CCDC78
|
coiled-coil domain containing 78
|
|
chr2_-_219633481
|
0.584
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr19_-_61874934
|
0.582
|
|
ZNF835
|
zinc finger protein 835
|
|
chr1_-_109853808
|
0.580
|
NM_020703
|
AMIGO1
|
adhesion molecule with Ig-like domain 1
|
|
chr6_-_10990048
|
0.578
|
NM_004752
|
GCM2
|
glial cells missing homolog 2 (Drosophila)
|
|
chr5_+_146594718
|
0.574
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr1_+_180836050
|
0.568
|
|
|
|
|
chr18_-_490583
|
0.567
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr4_-_11039600
|
0.564
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr19_-_14467939
|
0.551
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr3_+_46996150
|
0.547
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr8_-_143996257
|
0.543
|
NM_000498
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2
|
|
chr3_+_214668
|
0.541
|
|
|
|
|
chr6_+_41714171
|
0.541
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr14_+_28306633
|
0.531
|
|
FOXG1
|
forkhead box G1
|
|
chr1_+_12149662
|
0.531
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr3_-_18441763
|
0.521
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr19_-_41034734
|
0.520
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr19_-_10558906
|
0.516
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr19_+_60006877
|
0.516
|
NM_001080770
NM_001080772
NM_002255
|
KIR2DL4
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4
|
|
chr15_+_39032927
|
0.513
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr1_-_32054164
|
0.507
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr1_+_151917674
|
0.506
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr7_+_50314801
|
0.504
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr11_+_66381451
|
0.504
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chrX_-_139414890
|
0.503
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr8_-_29263714
|
0.500
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr8_-_26780645
|
0.500
|
|
|
|
|
chr19_+_40474867
|
0.497
|
NM_002361
NM_080600
|
MAG
|
myelin associated glycoprotein
|
|
chr1_+_11461764
|
0.496
|
NM_020780
|
PTCHD2
|
patched domain containing 2
|
|
chr6_-_166641860
|
0.495
|
NM_175922
|
PRR18
|
proline rich 18
|
|
chr17_+_1906262
|
0.494
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr7_-_131911808
|
0.494
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr14_+_101097440
|
0.492
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr19_-_50797258
|
0.486
|
|
GPR4
|
G protein-coupled receptor 4
|
|
chr2_+_130655970
|
0.486
|
NM_025029
|
MZT2B
|
mitotic spindle organizing protein 2B
|
|
chr7_-_143523582
|
0.485
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr1_-_231498081
|
0.479
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr1_+_25816545
|
0.473
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr17_+_1566566
|
0.473
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr19_-_10202947
|
0.471
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr4_-_647974
|
0.469
|
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta
|
|
chr8_+_23442307
|
0.464
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr12_-_6668680
|
0.463
|
|
ZNF384
|
zinc finger protein 384
|
|
chr16_+_65754788
|
0.463
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr5_+_31229609
|
0.462
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr17_+_37967946
|
0.461
|
|
|
|
|
chr7_+_72486044
|
0.461
|
NM_003508
|
FZD9
|
frizzled homolog 9 (Drosophila)
|
|
chr11_+_1367665
|
0.460
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr15_-_56093477
|
0.458
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr22_-_35970230
|
0.458
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr10_+_99248605
|
0.456
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr16_+_665680
|
0.451
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr11_-_407324
|
0.450
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr18_+_12244317
|
0.449
|
NM_001279
|
CIDEA
|
cell death-inducing DFFA-like effector a
|
|
chr19_-_55071645
|
0.437
|
|
AKT1S1
|
AKT1 substrate 1 (proline-rich)
|
|
chr19_-_43570503
|
0.437
|
NM_152657
|
GGN
|
gametogenetin
|
|
chr1_+_15941503
|
0.436
|
NM_001013641
|
TMEM82
|
transmembrane protein 82
|
|
chr16_-_1763036
|
0.431
|
NM_023936
|
MRPS34
|
mitochondrial ribosomal protein S34
|
|
chr21_-_41801950
|
0.428
|
NM_005656
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr19_-_18246166
|
0.426
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr3_-_173648896
|
0.425
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chr19_+_14108974
|
0.422
|
|
|
|
|
chr4_+_88939725
|
0.420
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr4_-_1232742
|
0.420
|
|
CTBP1
|
C-terminal binding protein 1
|
|
chr17_-_76064842
|
0.415
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr19_-_1125263
|
0.414
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chrX_-_100801472
|
0.412
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr1_-_200182338
|
0.412
|
NM_012134
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chrX_-_128616594
|
0.408
|
NM_017413
|
APLN
|
apelin
|
|
chr4_-_1232882
|
0.406
|
NM_001012614
NM_001328
|
CTBP1
|
C-terminal binding protein 1
|
|
chr11_+_66381077
|
0.406
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr4_-_123073642
|
0.403
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr16_-_17472238
|
0.401
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr17_-_70480385
|
0.400
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr10_-_48010879
|
0.398
|
NM_002900
|
RBP3
|
retinol binding protein 3, interstitial
|
|
chr19_+_43446937
|
0.396
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr11_-_117305277
|
0.396
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr7_-_131911768
|
0.393
|
|
PLXNA4
|
plexin A4
|
|
chr1_-_31942206
|
0.393
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr4_-_2390050
|
0.388
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr10_+_83625049
|
0.387
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr8_+_143542378
|
0.387
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|
|
chr11_+_128751044
|
0.386
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr22_-_37598160
|
0.385
|
|
CBX6
|
chromobox homolog 6
|
|
chr22_-_37598115
|
0.385
|
|
CBX6
|
chromobox homolog 6
|
|
chr7_+_143683365
|
0.385
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr19_-_5631502
|
0.385
|
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr16_+_87232501
|
0.381
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr12_+_111980044
|
0.381
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr9_-_130980359
|
0.376
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr13_-_37341859
|
0.374
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr17_-_71747867
|
0.371
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr16_+_23101538
|
0.371
|
NM_001039
|
SCNN1G
|
sodium channel, nonvoltage-gated 1, gamma
|
|
chr19_-_295778
|
0.368
|
NM_017550
|
MIER2
|
mesoderm induction early response 1, family member 2
|
|
chr8_-_57521702
|
0.368
|
NM_001135690
|
PENK
|
proenkephalin
|
|
chr1_-_31942142
|
0.368
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr4_+_94969064
|
0.367
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr11_-_67032125
|
0.367
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr1_+_154297588
|
0.366
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr3_-_193928038
|
0.366
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr2_-_197165495
|
0.365
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr19_-_1125257
|
0.364
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr8_-_105548323
|
0.364
|
NM_001385
|
DPYS
|
dihydropyrimidinase
|
|
chr9_-_109290107
|
0.364
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_+_38314837
|
0.363
|
NM_173479
|
WDR88
|
WD repeat domain 88
|
|
chr5_-_134942867
|
0.363
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr12_-_47679459
|
0.361
|
|
DDN
|
dendrin
|
|
chr11_+_63337421
|
0.360
|
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr11_+_65162114
|
0.358
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr7_-_143622188
|
0.357
|
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr6_-_40663089
|
0.357
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr10_+_72826656
|
0.356
|
NM_001171930
NM_001171931
NM_001171932
NM_022124
NM_052836
|
CDH23
|
cadherin-related 23
|
|
chr5_-_139992621
|
0.356
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr17_-_70220605
|
0.356
|
NM_139018
|
CD300LF
|
CD300 molecule-like family member f
|
|
chr22_-_18687511
|
0.354
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr5_+_76150637
|
0.354
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr18_+_41448763
|
0.353
|
NM_007163
|
SLC14A2
|
solute carrier family 14 (urea transporter), member 2
|
|
chr13_+_42046182
|
0.352
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr19_-_2259155
|
0.350
|
NM_001101391
|
LINGO3
|
leucine rich repeat and Ig domain containing 3
|
|
chr19_+_1358531
|
0.349
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr12_+_40117751
|
0.349
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr19_-_54093749
|
0.348
|
NM_003323
|
TULP2
|
tubby like protein 2
|
|
chr12_-_53064723
|
0.348
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr22_+_35777561
|
0.348
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr16_+_666075
|
0.346
|
NM_003961
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr16_-_1999796
|
0.345
|
|
ZNF598
|
zinc finger protein 598
|
|
chr11_+_66580861
|
0.345
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr9_-_130980302
|
0.344
|
|
IER5L
|
immediate early response 5-like
|
|
chr8_-_72918697
|
0.344
|
|
MSC
|
musculin
|
|
chr11_+_67567022
|
0.343
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr17_-_72045218
|
0.342
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr21_-_41140457
|
0.342
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr5_+_76150588
|
0.341
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr9_-_139202855
|
0.341
|
NM_013366
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr17_-_70480345
|
0.341
|
|
|
|
|
chr3_+_128874458
|
0.340
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr1_-_158883666
|
0.339
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr2_-_6924092
|
0.339
|
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr9_+_263025
|
0.339
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr1_-_200182090
|
0.339
|
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr6_+_16346759
|
0.338
|
NM_006877
|
GMPR
|
guanosine monophosphate reductase
|
|
chr22_+_39096576
|
0.338
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr11_+_63337498
|
0.337
|
NM_138471
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr16_-_1999760
|
0.337
|
NM_178167
|
ZNF598
|
zinc finger protein 598
|
|
chr7_+_156496360
|
0.337
|
|
LOC645249
|
hypothetical LOC645249
|
|
chr15_-_38332401
|
0.336
|
NM_001039905
|
C15orf56
|
chromosome 15 open reading frame 56
|
|
chr8_-_72918938
|
0.333
|
|
MSC
|
musculin
|
|
chr18_-_20231709
|
0.333
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr5_+_170779664
|
0.331
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr17_+_40192072
|
0.331
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr1_-_55039458
|
0.331
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chrX_+_48915127
|
0.330
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr20_+_2224612
|
0.329
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr12_+_121000938
|
0.329
|
|
MLXIP
|
MLX interacting protein
|
|
chr8_+_102573837
|
0.328
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_-_45081190
|
0.328
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr5_-_9599157
|
0.327
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr19_+_1358664
|
0.327
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr11_+_63337607
|
0.326
|
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr19_+_2240996
|
0.324
|
|
|
|
|
chr4_-_78038025
|
0.324
|
NM_001029870
|
ANKRD56
|
ankyrin repeat domain 56
|
|
chr11_+_64448628
|
0.324
|
NM_006244
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta
|
|
chr9_+_129951552
|
0.323
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr6_+_30002084
|
0.323
|
|
|
|
|
chrX_-_78509662
|
0.323
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr2_-_219566364
|
0.322
|
NM_005209
NM_057093
NM_057094
|
CRYBA2
|
crystallin, beta A2
|
|
chr8_+_32525269
|
0.322
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr2_-_133144094
|
0.321
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr22_+_39096563
|
0.320
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr3_-_153470022
|
0.320
|
|
LOC401093
|
hypothetical LOC401093
|