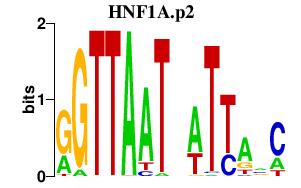
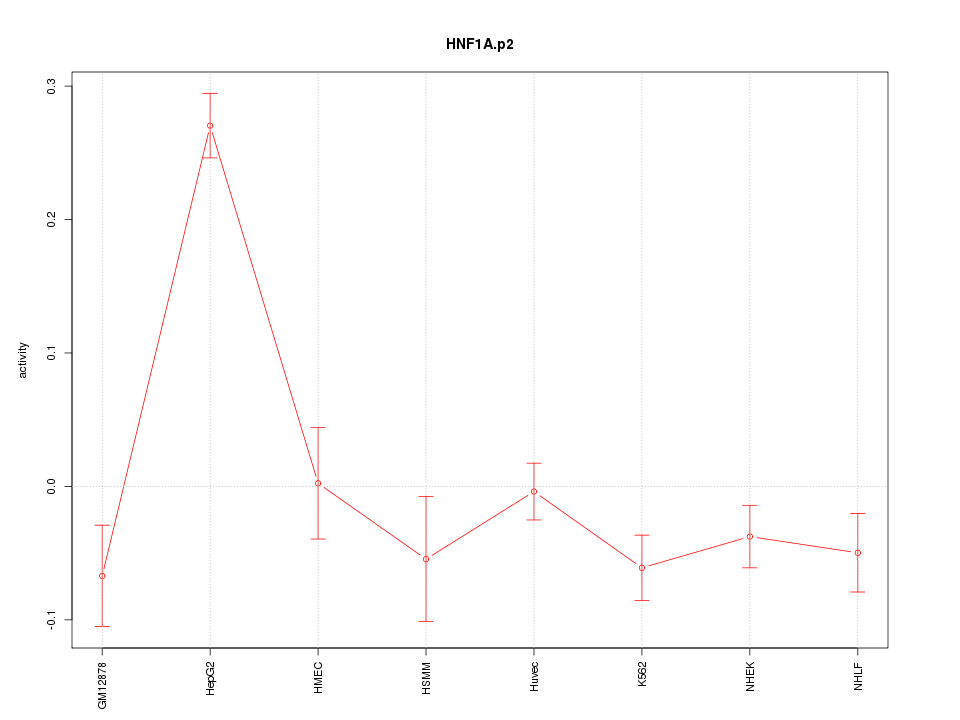
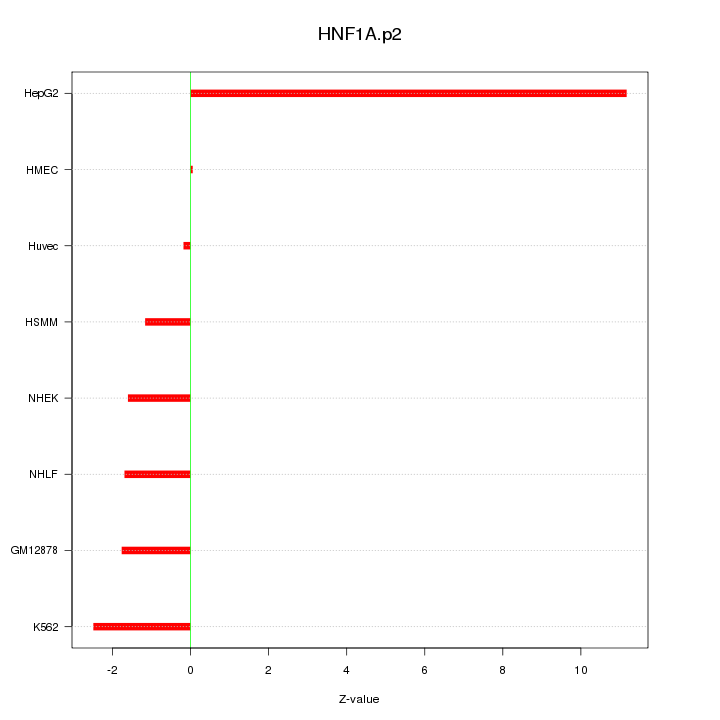
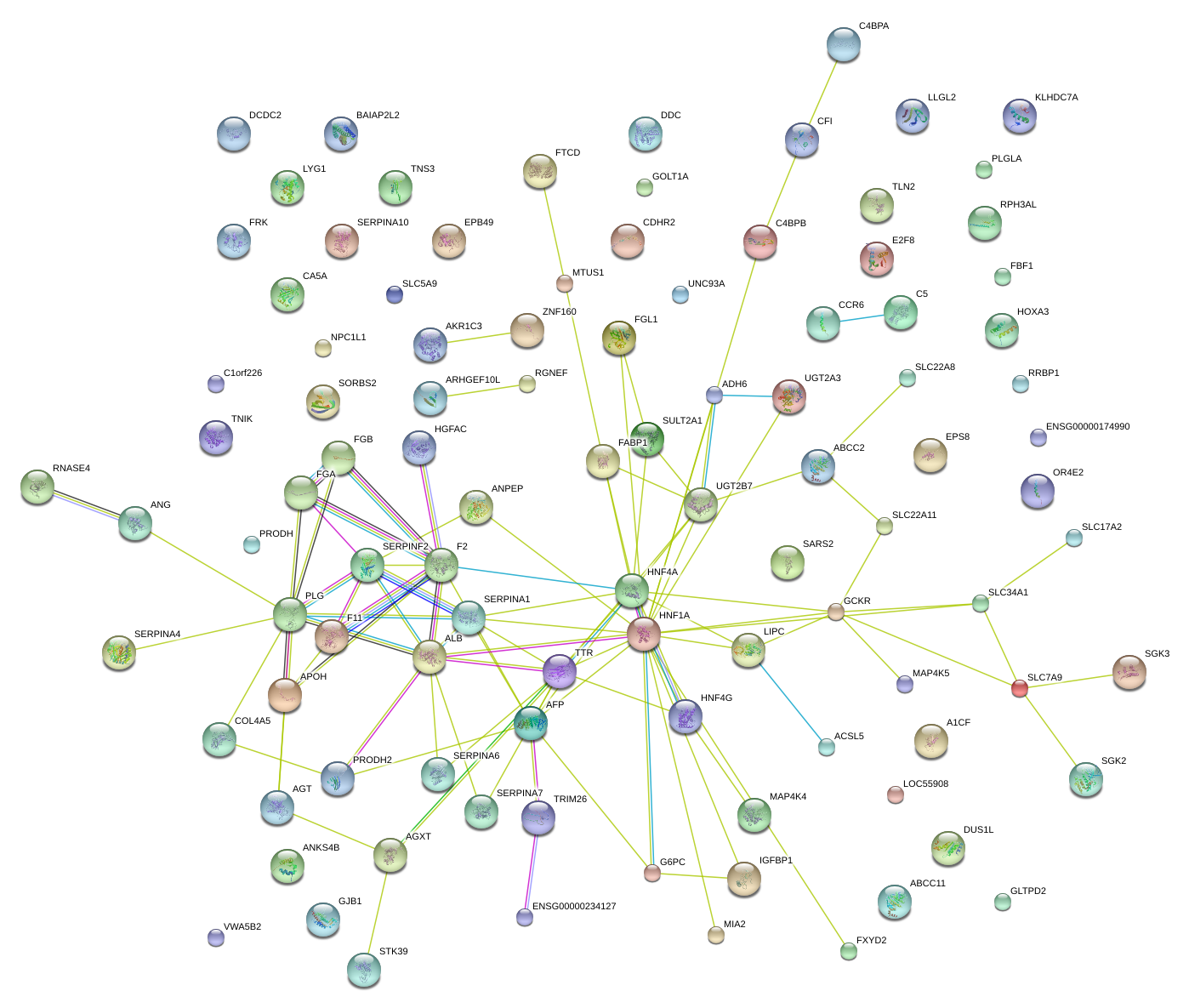
|
chr6_-_24491498
|
11.672
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chrX_-_105169370
|
10.675
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr17_-_61655915
|
8.959
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr20_+_42463279
|
8.913
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_+_42463350
|
8.030
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr14_-_93859394
|
7.901
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr1_+_48460943
|
7.488
|
NM_001011547
NM_001135181
|
SLC5A9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9
|
|
chr20_+_42417752
|
7.465
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr4_+_74520796
|
7.248
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr20_+_42463387
|
7.215
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr8_-_17797127
|
7.159
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr16_+_21152516
|
7.109
|
NM_145865
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr7_-_50600536
|
6.958
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr19_-_40995564
|
6.843
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr14_-_93924663
|
6.677
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr7_-_44547382
|
6.524
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr14_+_20226762
|
6.340
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr20_+_42463419
|
6.177
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr16_-_86527608
|
5.914
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr4_-_110942576
|
5.902
|
|
CFI
|
complement factor I
|
|
chr19_-_40995606
|
5.851
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr17_+_71065478
|
5.840
|
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr14_+_38772870
|
5.372
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr14_-_93924873
|
5.212
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr10_+_101532450
|
5.206
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr12_-_15757114
|
5.164
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr11_+_46697345
|
5.152
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr21_-_46399900
|
5.061
|
NM_006657
NM_206965
|
FTCD
|
formiminotransferase cyclodeaminase
|
|
chr2_+_27573206
|
4.972
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr4_-_110942783
|
4.960
|
NM_000204
|
CFI
|
complement factor I
|
|
chr4_-_155731288
|
4.862
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr14_-_93829309
|
4.789
|
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr17_+_38306339
|
4.751
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr4_+_74488835
|
4.749
|
NM_000477
|
ALB
|
albumin
|
|
chr11_+_46697312
|
4.738
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr9_-_122852356
|
4.720
|
NM_001735
|
C5
|
complement component 5
|
|
chr14_-_93829347
|
4.668
|
NM_001100607
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr8_-_17599438
|
4.469
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr14_+_94097513
|
4.459
|
NM_006215
|
SERPINA4
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4
|
|
chr4_-_69852090
|
4.378
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr10_-_52315390
|
4.332
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_-_93829113
|
4.302
|
NM_016186
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr5_+_175908948
|
4.148
|
NM_017675
|
CDHR2
|
cadherin-related family member 2
|
|
chr4_+_74488910
|
4.126
|
|
ALB
|
albumin
|
|
chr4_+_187424111
|
4.069
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr20_+_42417502
|
3.962
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr7_+_45894480
|
3.906
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr20_+_41621119
|
3.787
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr11_-_117200618
|
3.677
|
NM_001127489
NM_001680
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr4_+_74488947
|
3.651
|
|
ALB
|
albumin
|
|
chr4_-_100359353
|
3.624
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr10_+_5126589
|
3.516
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_101532544
|
3.496
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr10_-_52315436
|
3.415
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr15_-_88150823
|
3.393
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr10_+_114144512
|
3.318
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr20_+_41621079
|
3.251
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr17_+_1592879
|
3.213
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr1_+_17779528
|
3.199
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr8_+_21967008
|
3.182
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr10_+_5126613
|
3.172
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr17_+_4638966
|
3.041
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr6_-_26038817
|
2.910
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr19_-_53081326
|
2.906
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr1_+_93830106
|
2.890
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr10_+_5126556
|
2.865
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr2_-_88208650
|
2.829
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr19_-_53081383
|
2.812
|
NM_003167
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr6_+_167456230
|
2.719
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr7_-_47545717
|
2.602
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr1_+_205344200
|
2.505
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr22_-_36836478
|
2.491
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr8_+_76482661
|
2.482
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr15_+_56511466
|
2.481
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr4_+_3413489
|
2.404
|
NM_001528
|
HGFAC
|
HGF activator
|
|
chr4_+_3413457
|
2.391
|
|
HGFAC
|
HGF activator
|
|
chr4_+_69996780
|
2.364
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr4_+_187424336
|
2.351
|
|
F11
|
coagulation factor XI
|
|
chr17_-_202557
|
2.327
|
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr19_+_11211282
|
2.259
|
NM_018687
|
LOC55908
|
hepatocellular carcinoma-associated gene TD26
|
|
chr4_-_186970366
|
2.246
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_+_70351812
|
2.106
|
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr10_+_99334069
|
2.088
|
NM_001134670
NM_138413
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr17_-_202617
|
2.083
|
NM_001190411
NM_001190412
NM_001190413
NM_006987
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr17_+_1592893
|
2.071
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr2_+_101874765
|
2.066
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chrX_+_70351773
|
2.026
|
NM_001097642
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr16_-_46826588
|
1.988
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr6_-_116488458
|
1.963
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr18_+_27425727
|
1.948
|
NM_000371
|
TTR
|
transthyretin
|
|
chr15_+_60726801
|
1.945
|
NM_015059
|
TLN2
|
talin 2
|
|
chr19_-_38052481
|
1.884
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr11_-_62539857
|
1.879
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr1_-_202449659
|
1.878
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr4_+_155703581
|
1.876
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr4_+_74497998
|
1.857
|
|
ALB
|
albumin
|
|
chr1_+_205328834
|
1.807
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr7_-_27133163
|
1.792
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr2_-_99284070
|
1.774
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr11_+_64079665
|
1.754
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr6_+_161043214
|
1.752
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr2_-_168812132
|
1.719
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr5_+_73145098
|
1.713
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr2_+_101874809
|
1.676
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr6_+_167624792
|
1.676
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr1_+_160618143
|
1.662
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr17_+_1593040
|
1.631
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr3_+_185430910
|
1.583
|
NM_138345
|
VWA5B2
|
von Willebrand factor A domain containing 5B2
|
|
chr1_-_228916559
|
1.564
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr4_+_74498042
|
1.536
|
|
ALB
|
albumin
|
|
chr14_+_21203136
|
1.522
|
NM_001001912
|
OR4E2
|
olfactory receptor, family 4, subfamily E, member 2
|
|
chr20_-_17589122
|
1.502
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr19_-_44135166
|
1.491
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr1_+_18680010
|
1.443
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr19_-_40996040
|
1.439
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chrX_+_107569671
|
1.417
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr12_+_119900677
|
1.411
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr4_-_70115037
|
1.378
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr11_+_46697335
|
1.305
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr7_+_94130673
|
1.272
|
|
PEG10
|
paternally expressed 10
|
|
chr3_-_168674502
|
1.269
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr1_+_93830169
|
1.268
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr14_+_38805251
|
1.241
|
NM_203356
|
CTAGE5
|
CTAGE family, member 5
|
|
chr4_+_56731117
|
1.233
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chr1_-_157551072
|
1.218
|
NM_001004467
|
OR10J3
|
olfactory receptor, family 10, subfamily J, member 3
|
|
chr1_+_81544432
|
1.183
|
|
LPHN2
|
latrophilin 2
|
|
chr1_+_157824239
|
1.150
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr4_-_69116839
|
1.122
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr2_-_207291364
|
1.114
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr1_-_20014180
|
1.108
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chr16_-_20271530
|
1.097
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr5_+_176446425
|
1.091
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr20_+_61841553
|
1.077
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr7_+_72913424
|
1.075
|
NM_182504
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr5_-_147191428
|
1.040
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr8_+_98772537
|
1.026
|
|
MTDH
|
metadherin
|
|
chr2_-_63518589
|
1.018
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr5_-_147191334
|
1.004
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr14_+_54238581
|
0.988
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr7_+_94953148
|
0.986
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr2_+_234191029
|
0.977
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr7_-_156494741
|
0.947
|
NM_001165255
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr16_-_56393871
|
0.945
|
|
KIFC3
|
kinesin family member C3
|
|
chr2_+_28572441
|
0.923
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr9_-_115901157
|
0.896
|
NM_138424
|
KIF12
|
kinesin family member 12
|
|
chr17_+_77910411
|
0.883
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr6_-_25982418
|
0.864
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr2_+_234302511
|
0.863
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr2_+_62787534
|
0.855
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr10_-_121592020
|
0.844
|
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr17_+_65009986
|
0.831
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr6_-_52736231
|
0.815
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr6_+_158653679
|
0.796
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr5_+_76470829
|
0.788
|
|
|
|
|
chr3_-_162572441
|
0.755
|
NM_001040100
|
C3orf57
|
chromosome 3 open reading frame 57
|
|
chr20_+_55569542
|
0.754
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr1_-_228916486
|
0.754
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr18_-_44723116
|
0.725
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr7_-_107230867
|
0.720
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chrX_-_131375217
|
0.719
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_+_64289585
|
0.715
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr16_+_20682812
|
0.699
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr14_+_64240996
|
0.694
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr6_-_133161318
|
0.685
|
NM_052831
|
C6orf192
|
chromosome 6 open reading frame 192
|
|
chr4_-_159312973
|
0.685
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr22_-_36836620
|
0.684
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr19_+_50109416
|
0.682
|
|
APOC1
|
apolipoprotein C-I
|
|
chr13_-_32658215
|
0.681
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_+_74493918
|
0.657
|
|
ALB
|
albumin
|
|
chr4_+_159350850
|
0.650
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr3_+_12367993
|
0.641
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr1_-_228916470
|
0.638
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr20_+_2892918
|
0.638
|
|
PTPRA
|
protein tyrosine phosphatase, receptor type, A
|
|
chr12_-_12182641
|
0.625
|
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr16_+_88224175
|
0.620
|
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr20_-_43972061
|
0.617
|
|
PLTP
|
phospholipid transfer protein
|
|
chr5_-_160044844
|
0.614
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr8_-_18585721
|
0.609
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr14_+_54291255
|
0.608
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr2_+_101981888
|
0.608
|
NM_173343
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr14_+_67238355
|
0.605
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr3_+_187866464
|
0.595
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr22_-_41685827
|
0.595
|
NM_001184971
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr2_+_10103735
|
0.594
|
|
|
|
|
chr12_-_8584658
|
0.590
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr6_+_131613196
|
0.587
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr3_+_189425886
|
0.578
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_101398288
|
0.573
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_157950898
|
0.571
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr6_-_29563657
|
0.558
|
NM_052967
|
MAS1L
|
MAS1 oncogene-like
|
|
chr11_+_64114857
|
0.557
|
NM_144585
NM_153378
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12
|
|
chr6_-_119512035
|
0.535
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr12_+_10222823
|
0.532
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr16_+_88224276
|
0.530
|
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr21_-_35153664
|
0.524
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_+_79829794
|
0.513
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr4_+_124537334
|
0.503
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_145892190
|
0.501
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr1_-_195818927
|
0.498
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr3_+_191588354
|
0.495
|
NM_006580
|
CLDN16
|
claudin 16
|