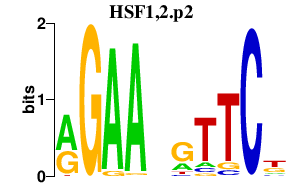
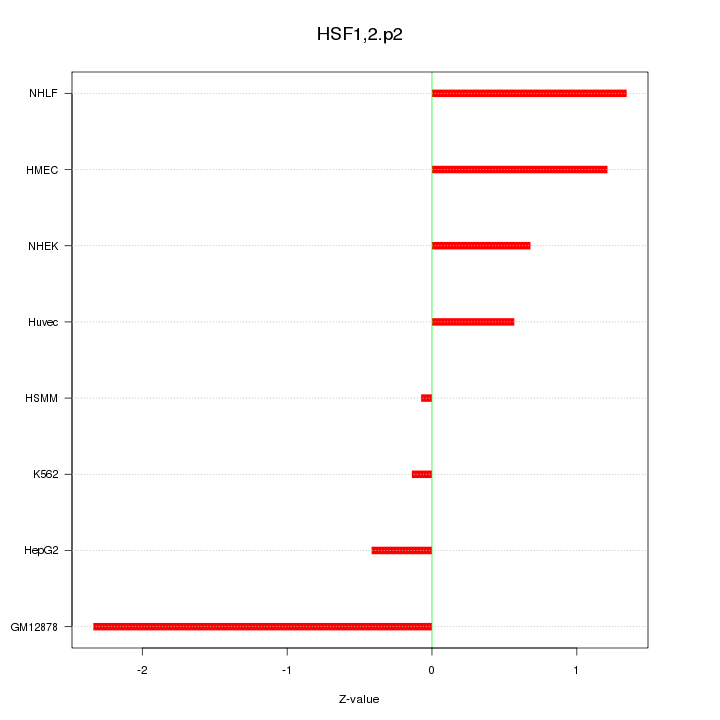
|
chr6_+_44234525
|
1.710
|
NM_007058
|
CAPN11
|
calpain 11
|
|
chr12_+_118100977
|
1.552
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr5_-_110090264
|
1.466
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr3_-_36756355
|
1.453
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chr5_-_112798436
|
1.331
|
NM_032028
|
TSSK1B
|
testis-specific serine kinase 1B
|
|
chr11_+_12265022
|
1.288
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr6_-_31890765
|
1.248
|
NM_005527
|
HSPA1L
|
heat shock 70kDa protein 1-like
|
|
chr9_-_90983501
|
1.230
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr2_+_210152609
|
1.225
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_+_12757570
|
1.200
|
NM_001080830
|
PRAMEF12
|
PRAME family member 12
|
|
chr19_+_40298571
|
1.180
|
NM_001136007
NM_001136008
NM_001136009
NM_001136010
NM_001136011
NM_001136012
NM_005971
NM_021910
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr14_-_21108714
|
1.157
|
NM_001005465
|
OR10G3
|
olfactory receptor, family 10, subfamily G, member 3
|
|
chr12_-_12994584
|
1.096
|
NM_018654
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D
|
|
chr20_+_31334601
|
1.092
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr11_+_68818175
|
1.077
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr7_-_75876830
|
1.074
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr6_+_54281161
|
1.036
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr22_+_22907443
|
1.035
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr6_+_29472310
|
1.006
|
NM_013936
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2
|
|
chr2_-_113259441
|
0.990
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr11_-_104411065
|
0.984
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr6_+_31891259
|
0.980
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr17_-_36890877
|
0.973
|
NM_002280
|
KRT35
|
keratin 35
|
|
chr9_+_108726257
|
0.959
|
|
ZNF462
|
zinc finger protein 462
|
|
chr7_-_27133163
|
0.953
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr3_-_64648703
|
0.949
|
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr7_-_80386258
|
0.947
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr4_+_111053488
|
0.942
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr1_-_173979292
|
0.941
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr5_+_79366746
|
0.917
|
NM_003248
|
THBS4
|
thrombospondin 4
|
|
chr1_+_155052094
|
0.910
|
NM_001007792
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1
|
|
chr5_+_147423727
|
0.893
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr7_-_80386602
|
0.889
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr2_-_217268492
|
0.886
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_+_32698892
|
0.885
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr12_+_8557402
|
0.879
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr2_-_197383120
|
0.878
|
NM_213608
|
C2orf66
|
chromosome 2 open reading frame 66
|
|
chr12_-_14887648
|
0.870
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr12_+_47584159
|
0.864
|
NM_033124
|
CCDC65
|
coiled-coil domain containing 65
|
|
chr6_+_31891293
|
0.862
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr7_-_36730525
|
0.862
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr6_+_31190505
|
0.849
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr5_-_41107161
|
0.830
|
NM_173489
|
HEATR7B2
|
HEAT repeat family member 7B2
|
|
chr20_+_55338221
|
0.823
|
NM_012444
NM_198265
|
SPO11
|
SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae)
|
|
chr19_+_50693570
|
0.820
|
NM_001080401
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative)
|
|
chr10_+_122206455
|
0.819
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr5_-_135318420
|
0.797
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr17_+_53670785
|
0.795
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr2_+_219532590
|
0.794
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr5_-_22248630
|
0.792
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr15_+_52092392
|
0.792
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr7_-_127042810
|
0.785
|
NM_006193
|
PAX4
|
paired box 4
|
|
chr2_+_161981138
|
0.779
|
|
TBR1
|
T-box, brain, 1
|
|
chr14_+_62740878
|
0.778
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr16_+_4546491
|
0.773
|
NM_001145011
|
LOC342346
|
hypothetical protein LOC342346
|
|
chr10_+_18280773
|
0.751
|
NM_001145195
NM_152725
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chr5_-_148422799
|
0.750
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr22_+_22907417
|
0.746
|
|
SUSD2
|
sushi domain containing 2
|
|
chr2_+_166137091
|
0.730
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_+_160269734
|
0.729
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr1_-_148175383
|
0.728
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr12_-_3472245
|
0.726
|
|
LOC100129223
|
hypothetical LOC100129223
|
|
chr12_+_120055013
|
0.725
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr7_+_129807467
|
0.723
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr12_-_121753795
|
0.721
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr19_-_54258935
|
0.719
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr2_-_142605739
|
0.717
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr2_+_26768894
|
0.716
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr1_-_112333192
|
0.712
|
NM_004980
NM_172198
|
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3
|
|
chr1_-_150353179
|
0.704
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr11_-_34491878
|
0.702
|
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr20_+_19818209
|
0.702
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_-_22458134
|
0.698
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr2_+_234292162
|
0.698
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr6_+_31903374
|
0.694
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr11_-_10671736
|
0.694
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr3_-_9265592
|
0.691
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chrX_+_152454773
|
0.688
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr11_-_6298284
|
0.688
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr4_+_77575276
|
0.687
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr6_+_46728590
|
0.683
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr8_-_37916803
|
0.683
|
NM_152413
|
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1
|
|
chr20_-_61574259
|
0.680
|
NM_004518
NM_172106
NM_172107
NM_172108
NM_172109
|
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2
|
|
chr1_+_47674275
|
0.679
|
NM_004474
|
FOXD2
|
forkhead box D2
|
|
chr18_+_27152049
|
0.677
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr19_-_54252813
|
0.664
|
|
CGB7
|
chorionic gonadotropin, beta polypeptide 7
|
|
chr9_+_83748234
|
0.664
|
NM_207416
|
FAM75D3
|
family with sequence similarity 75, member D3
|
|
chr10_-_90601655
|
0.663
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr17_+_21249040
|
0.661
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr19_-_60557875
|
0.660
|
NM_144613
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis)
|
|
chr10_+_135190855
|
0.657
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr4_-_153820535
|
0.655
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr2_-_189752575
|
0.655
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr17_-_24068759
|
0.645
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr11_-_35503720
|
0.637
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr1_-_200946038
|
0.636
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr6_+_32817140
|
0.629
|
NM_020056
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2
|
|
chr16_+_54158084
|
0.623
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr1_+_109450116
|
0.620
|
|
|
|
|
chr9_-_89939719
|
0.614
|
NM_001166137
|
FAM75C2
|
family with sequence similarity 75, member C2
|
|
chr11_+_1812115
|
0.614
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr6_+_116939500
|
0.608
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr9_+_831689
|
0.606
|
NM_021951
|
DMRT1
|
doublesex and mab-3 related transcription factor 1
|
|
chr15_-_39973566
|
0.605
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr19_+_3073297
|
0.605
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr1_-_158190585
|
0.599
|
NM_001146172
NM_001146173
NM_033438
|
SLAMF9
|
SLAM family member 9
|
|
chr12_-_51253875
|
0.598
|
NM_175053
|
KRT74
|
keratin 74
|
|
chr3_-_48607596
|
0.598
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr19_-_53941835
|
0.597
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr2_+_202379442
|
0.596
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr18_+_18003401
|
0.591
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr16_-_73012740
|
0.590
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr17_-_76897590
|
0.588
|
NM_178519
|
C17orf55
|
chromosome 17 open reading frame 55
|
|
chr7_-_83662152
|
0.586
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr12_-_21818881
|
0.584
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr11_-_47692877
|
0.581
|
NM_024783
|
AGBL2
|
ATP/GTP binding protein-like 2
|
|
chr2_-_39040981
|
0.580
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr8_+_22078622
|
0.578
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr19_+_3073560
|
0.578
|
|
|
|
|
chr3_+_44665220
|
0.577
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr6_+_134800533
|
0.576
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr2_+_220007943
|
0.573
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr2_+_210152399
|
0.572
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr12_+_10054492
|
0.571
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chr1_-_148175345
|
0.566
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr4_-_160175782
|
0.566
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr11_-_121492010
|
0.563
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr12_-_121224676
|
0.563
|
NM_001014336
|
IL31
|
interleukin 31
|
|
chr5_-_146813343
|
0.563
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr6_+_152168500
|
0.562
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr14_-_22458199
|
0.561
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr7_+_75769927
|
0.560
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr3_-_74652952
|
0.560
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr11_-_10671608
|
0.557
|
NM_001100167
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr19_-_60383280
|
0.556
|
NM_003180
|
SYT5
|
synaptotagmin V
|
|
chr9_-_90983195
|
0.552
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr19_-_14750347
|
0.552
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr6_+_83129641
|
0.552
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr17_-_71648965
|
0.550
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr8_-_142307854
|
0.543
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr4_-_123092358
|
0.541
|
NM_001130698
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr3_-_135131207
|
0.540
|
NM_025041
|
C3orf36
|
chromosome 3 open reading frame 36
|
|
chr2_-_169596078
|
0.538
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr17_-_10217046
|
0.536
|
NM_003802
|
MYH13
|
myosin, heavy chain 13, skeletal muscle
|
|
chr12_-_83830710
|
0.535
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr3_+_191588354
|
0.535
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr8_+_39890484
|
0.534
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr19_+_15079141
|
0.531
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr12_-_121767272
|
0.531
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr9_-_116732403
|
0.526
|
NM_001244
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8
|
|
chr11_+_89458765
|
0.525
|
NM_001143975
|
UBTFL1
|
upstream binding transcription factor, RNA polymerase I-like 1
|
|
chr3_+_1109619
|
0.525
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chrX_-_10495476
|
0.523
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr9_-_72926333
|
0.522
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr3_-_184756101
|
0.522
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr3_+_98016114
|
0.520
|
NM_001080448
|
EPHA6
|
EPH receptor A6
|
|
chr11_+_64079665
|
0.518
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr2_+_113533155
|
0.518
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr19_-_14490227
|
0.516
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr6_-_169392771
|
0.514
|
|
|
|
|
chr9_+_36159384
|
0.514
|
NM_005893
|
CCIN
|
calicin
|
|
chr7_-_4867963
|
0.513
|
NM_020144
|
PAPOLB
|
poly(A) polymerase beta (testis specific)
|
|
chr3_-_64186151
|
0.513
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr14_-_76564374
|
0.509
|
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr17_+_7199178
|
0.509
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr12_-_51298599
|
0.509
|
NM_175068
|
KRT73
|
keratin 73
|
|
chrX_-_110542040
|
0.507
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr6_+_47774247
|
0.506
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr8_-_26427304
|
0.505
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr18_-_26996742
|
0.503
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr14_-_59407103
|
0.502
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr5_-_22889487
|
0.500
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr6_-_117193505
|
0.500
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr8_-_23768242
|
0.498
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr22_+_30769018
|
0.496
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr17_-_40348378
|
0.496
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr11_-_27637771
|
0.496
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_159510416
|
0.494
|
NM_032532
|
FNDC1
|
fibronectin type III domain containing 1
|
|
chr12_+_6768966
|
0.491
|
|
CD4
|
CD4 molecule
|
|
chr2_+_234490781
|
0.489
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr1_-_150092702
|
0.489
|
NM_182578
|
THEM5
|
thioesterase superfamily member 5
|
|
chr5_+_140762551
|
0.488
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr19_+_56049982
|
0.488
|
NM_001030047
NM_001030048
NM_001030050
NM_001648
|
KLK3
|
kallikrein-related peptidase 3
|
|
chr3_-_59010777
|
0.482
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr7_+_134114912
|
0.480
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_83052606
|
0.479
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr5_+_139485703
|
0.477
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr4_+_71713324
|
0.477
|
NM_031889
|
ENAM
|
enamelin
|
|
chr14_+_28306633
|
0.474
|
|
FOXG1
|
forkhead box G1
|
|
chr16_+_29697090
|
0.472
|
|
ZG16
|
zymogen granule protein 16 homolog (rat)
|
|
chr11_-_133332089
|
0.471
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr9_+_34979674
|
0.468
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr8_-_68821131
|
0.468
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr13_+_109932934
|
0.465
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr18_+_29573152
|
0.465
|
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr22_+_44828338
|
0.462
|
|
|
|
|
chr7_-_14847479
|
0.461
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr14_-_80934523
|
0.461
|
NM_033104
|
STON2
|
stonin 2
|
|
chr5_-_180560535
|
0.458
|
NM_203297
|
TRIM7
|
tripartite motif containing 7
|
|
chr11_+_123405506
|
0.457
|
NM_001004464
|
OR10G8
|
olfactory receptor, family 10, subfamily G, member 8
|