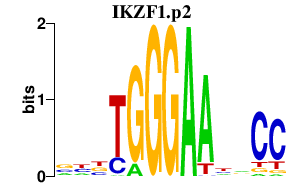
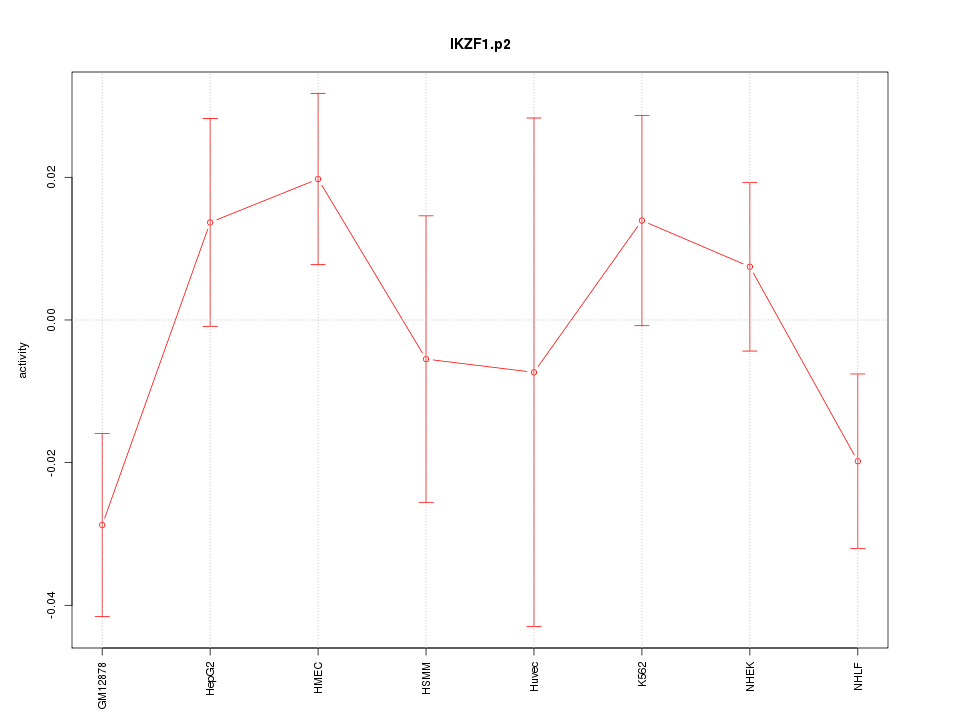
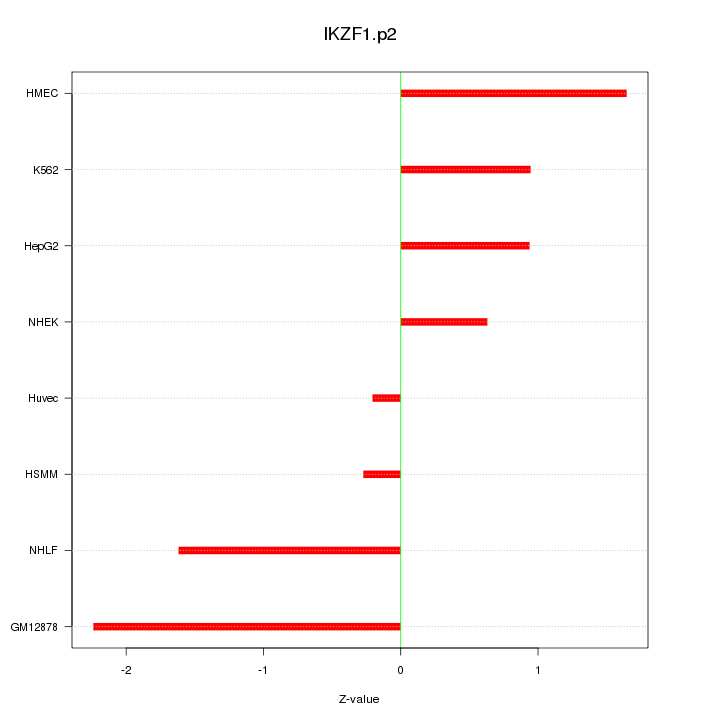
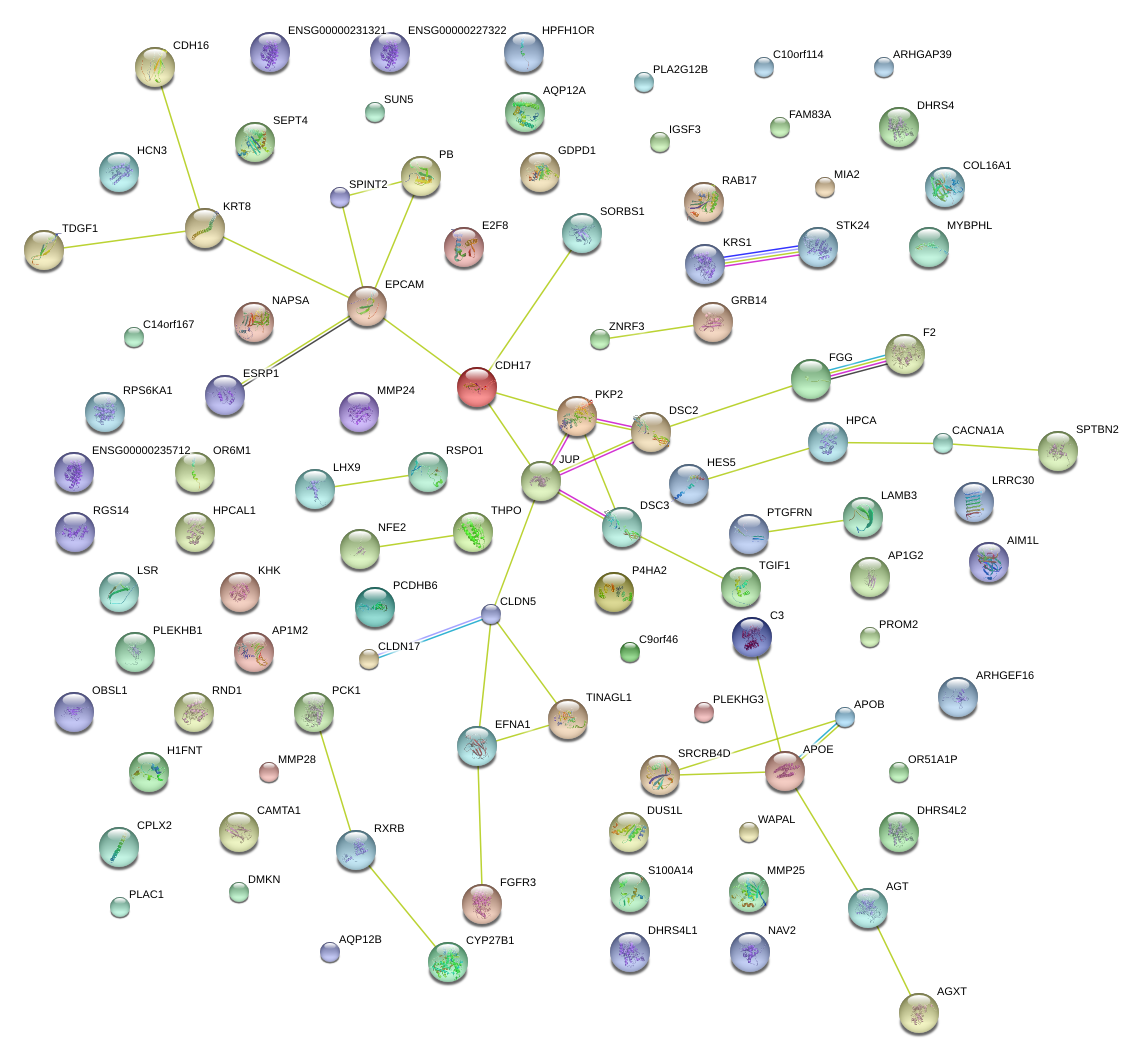
|
chr12_-_47545816
|
1.206
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr20_+_55569542
|
1.102
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr19_-_10558906
|
0.993
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr1_+_117253882
|
0.885
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr14_+_38772870
|
0.800
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr6_+_132901119
|
0.671
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr21_-_30460841
|
0.668
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr2_-_238164452
|
0.660
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr1_+_3378030
|
0.634
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr1_-_109651140
|
0.631
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr3_-_185578621
|
0.630
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr4_-_155753207
|
0.625
|
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_124263932
|
0.602
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr1_+_6767968
|
0.600
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr1_+_26728846
|
0.599
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr4_-_155753332
|
0.591
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr1_+_26728827
|
0.583
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr2_+_47449790
|
0.576
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr22_-_17892859
|
0.569
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr1_+_26728839
|
0.567
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr1_+_33124658
|
0.565
|
NM_002143
|
HPCA
|
hippocalcin
|
|
chr2_-_220144254
|
0.560
|
|
OBSL1
|
obscurin-like 1
|
|
chr12_-_56447090
|
0.557
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr2_-_21120398
|
0.553
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_+_31814712
|
0.551
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr8_-_145881968
|
0.548
|
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr11_-_123182266
|
0.547
|
NM_001005325
|
OR6M1
|
olfactory receptor, family 6, subfamily M, member 1
|
|
chr22_+_27609889
|
0.545
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr7_-_142695392
|
0.545
|
|
|
|
|
chr3_+_46594079
|
0.543
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr1_-_228916486
|
0.536
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr4_-_155753249
|
0.535
|
|
FGG
|
fibrinogen gamma chain
|
|
chr4_-_155753287
|
0.524
|
|
FGG
|
fibrinogen gamma chain
|
|
chr1_-_2451543
|
0.524
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr2_+_241279934
|
0.523
|
NM_198998
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr14_-_23106779
|
0.519
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr12_-_51585004
|
0.518
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr1_-_117011824
|
0.516
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr4_-_155753272
|
0.512
|
|
FGG
|
fibrinogen gamma chain
|
|
chr19_-_6643953
|
0.511
|
|
C3
|
complement component 3
|
|
chr17_+_54652609
|
0.509
|
NM_001165993
NM_001165994
NM_182569
|
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1
|
|
chr1_-_228916470
|
0.508
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr2_+_95303947
|
0.507
|
|
PROM2
|
prominin 2
|
|
chr2_-_238164042
|
0.498
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr17_+_77478858
|
0.497
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr18_+_3441589
|
0.493
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr10_-_97190885
|
0.487
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr3_-_44464722
|
0.484
|
|
|
|
|
chr2_-_241270982
|
0.479
|
NM_001102467
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr1_+_153366559
|
0.477
|
|
EFNA1
|
ephrin-A1
|
|
chr1_+_31814709
|
0.473
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chrX_-_133620103
|
0.473
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr1_+_153513810
|
0.471
|
NM_020897
|
HCN3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3
|
|
chr10_-_21825879
|
0.470
|
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr1_-_228916559
|
0.468
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr7_-_75876830
|
0.468
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr11_-_5130174
|
0.465
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr2_-_21120314
|
0.457
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr18_+_7221136
|
0.453
|
NM_001105581
|
LRRC30
|
leucine rich repeat containing 30
|
|
chr1_-_207892420
|
0.452
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr5_+_175231098
|
0.444
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr1_-_31942206
|
0.443
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr1_-_26553168
|
0.442
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr14_-_20642214
|
0.440
|
|
|
|
|
chr5_+_176726918
|
0.436
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr16_+_3036682
|
0.432
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr12_-_32940931
|
0.429
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr12_-_52980984
|
0.429
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr11_-_66245288
|
0.427
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr1_+_31814672
|
0.426
|
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr11_+_73034870
|
0.424
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr12_-_52980914
|
0.422
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr2_+_95303922
|
0.418
|
NM_001165977
NM_001165978
NM_144707
|
PROM2
|
prominin 2
|
|
chrX_+_130984912
|
0.418
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr14_+_23527866
|
0.417
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr11_+_46697345
|
0.415
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr6_-_33276416
|
0.414
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr19_-_40693176
|
0.414
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr17_-_37181636
|
0.413
|
|
JUP
|
junction plakoglobin
|
|
chr14_-_23107106
|
0.413
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr1_-_37872874
|
0.406
|
|
RSPO1
|
R-spondin homolog (Xenopus laevis)
|
|
chr20_-_31055892
|
0.402
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr12_+_47009029
|
0.402
|
NM_181788
|
H1FNT
|
H1 histone family, member N, testis-specific
|
|
chr17_-_53973177
|
0.401
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr2_+_27163114
|
0.398
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr9_-_5620070
|
0.391
|
|
C9orf46
|
chromosome 9 open reading frame 46
|
|
chr1_-_151855413
|
0.385
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr8_+_95722516
|
0.385
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_-_207892296
|
0.384
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr2_-_165133239
|
0.383
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr4_+_1764802
|
0.382
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr7_-_96471544
|
0.382
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr14_-_23493169
|
0.374
|
|
C14orf167
|
chromosome 14 open reading frame 167
|
|
chr18_-_26935883
|
0.373
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr10_-_74384569
|
0.372
|
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr1_+_196153122
|
0.371
|
NM_020204
|
LHX9
|
LIM homeobox 9
|
|
chr3_-_12561962
|
0.370
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr19_+_40431122
|
0.370
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr16_-_65510230
|
0.367
|
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr5_+_140509803
|
0.365
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr10_-_21826192
|
0.363
|
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr14_+_64240996
|
0.362
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr10_-_21826211
|
0.362
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr5_-_131591380
|
0.361
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr11_+_20000938
|
0.360
|
|
NAV2
|
neuron navigator 2
|
|
chr19_+_43446937
|
0.358
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr14_+_64240902
|
0.357
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr7_-_103417198
|
0.349
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr1_+_203740391
|
0.348
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr10_-_74384485
|
0.347
|
NM_032562
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr6_+_31190505
|
0.346
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr7_-_99536852
|
0.345
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr19_+_43447262
|
0.342
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_43447078
|
0.341
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr8_+_95722575
|
0.341
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_+_15123165
|
0.338
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr6_+_35856798
|
0.338
|
NM_001010886
|
C6orf127
|
chromosome 6 open reading frame 127
|
|
chr6_-_30188845
|
0.335
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr19_-_56147971
|
0.334
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr17_+_53769659
|
0.332
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr11_-_18214891
|
0.331
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr1_-_95472096
|
0.330
|
|
|
|
|
chr20_+_41621119
|
0.327
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr19_+_54314292
|
0.322
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr11_+_384199
|
0.320
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr9_+_129951552
|
0.320
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr6_-_109521969
|
0.320
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr19_+_50004134
|
0.319
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr18_+_3443771
|
0.319
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr20_+_41621079
|
0.317
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr16_+_3102556
|
0.317
|
NM_001042428
NM_003456
|
ZNF205
|
zinc finger protein 205
|
|
chr17_+_23818922
|
0.313
|
|
|
|
|
chr1_+_207915282
|
0.307
|
NM_015714
|
G0S2
|
G0/G1switch 2
|
|
chr3_+_46423682
|
0.306
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr17_+_50333021
|
0.302
|
NM_005486
|
TOM1L1
|
target of myb1 (chicken)-like 1
|
|
chr5_+_109052984
|
0.302
|
|
|
|
|
chr1_+_160599396
|
0.301
|
NM_001126060
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr16_-_69277194
|
0.301
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr19_-_17217022
|
0.299
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr16_+_2742684
|
0.296
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr22_+_18447732
|
0.285
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chr17_-_37181626
|
0.283
|
|
|
|
|
chr1_+_32992131
|
0.283
|
|
|
|
|
chr6_-_2696152
|
0.281
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr8_-_17787342
|
0.280
|
|
FGL1
|
fibrinogen-like 1
|
|
chr4_+_128773529
|
0.276
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr2_+_96295715
|
0.275
|
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1
|
|
chr14_-_68424653
|
0.274
|
|
ACTN1
|
actinin, alpha 1
|
|
chr22_-_28492933
|
0.274
|
NM_001003692
NM_019103
|
ZMAT5
|
zinc finger, matrin-type 5
|
|
chr7_-_50600536
|
0.273
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr22_+_40347263
|
0.273
|
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr1_+_12627152
|
0.272
|
NM_001013630
|
AADACL4
|
arylacetamide deacetylase-like 4
|
|
chr8_+_95722448
|
0.271
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr12_+_50913220
|
0.269
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr6_+_150505849
|
0.269
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr17_+_7457988
|
0.269
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr8_-_7718769
|
0.268
|
NM_152250
NM_001040703
|
DEFB105A
DEFB105B
|
defensin, beta 105A
defensin, beta 105B
|
|
chr2_-_110665188
|
0.268
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr17_+_53769562
|
0.267
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr17_+_69781980
|
0.267
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr2_-_21081694
|
0.265
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr3_-_140148671
|
0.265
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr5_-_150441150
|
0.264
|
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr1_-_232962369
|
0.262
|
|
PP2672
|
hypothetical LOC100130249
|
|
chr21_+_44534575
|
0.261
|
NM_000658
|
AIRE
|
autoimmune regulator
|
|
chr3_+_195336631
|
0.257
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr11_-_16992524
|
0.255
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr19_+_59062962
|
0.254
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_-_208046011
|
0.254
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr11_+_5466490
|
0.254
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr17_-_58877276
|
0.253
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr1_-_110108048
|
0.253
|
NM_024526
NM_133181
NM_139053
|
EPS8L3
|
EPS8-like 3
|
|
chr2_+_17585462
|
0.252
|
|
VSNL1
|
visinin-like 1
|
|
chr11_+_6216905
|
0.250
|
NM_001037329
|
CNGA4
|
cyclic nucleotide gated channel alpha 4
|
|
chr2_+_17585287
|
0.250
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr10_-_52315390
|
0.249
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr19_-_50779914
|
0.248
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr8_+_38007147
|
0.248
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr19_-_50618471
|
0.248
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr20_+_58064360
|
0.248
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr17_+_24079924
|
0.247
|
NM_178170
|
NEK8
|
NIMA (never in mitosis gene a)- related kinase 8
|
|
chr5_+_137447479
|
0.246
|
NM_058244
|
WNT8A
|
wingless-type MMTV integration site family, member 8A
|
|
chr1_+_168381811
|
0.246
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr14_+_22845851
|
0.246
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr17_-_31882135
|
0.246
|
NM_001001418
NM_001040282
|
TBC1D3C
TBC1D3G
TBC1D3
|
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3
|
|
chr2_+_28572441
|
0.245
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr17_-_77411707
|
0.244
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr1_+_35020425
|
0.243
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr22_+_38720881
|
0.242
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr5_+_66160359
|
0.242
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr14_+_22633728
|
0.242
|
|
C14orf119
|
chromosome 14 open reading frame 119
|
|
chr6_-_15356865
|
0.241
|
|
|
|
|
chr1_-_153514124
|
0.241
|
|
CLK2
|
CDC-like kinase 2
|
|
chr11_+_22646229
|
0.240
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr6_-_47117951
|
0.240
|
NM_025048
NM_153840
|
GPR110
|
G protein-coupled receptor 110
|
|
chr2_-_227372574
|
0.240
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr6_-_31812049
|
0.239
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr17_-_77411659
|
0.239
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr6_-_31812214
|
0.239
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr20_-_1113116
|
0.239
|
NM_018354
|
C20orf46
|
chromosome 20 open reading frame 46
|