|
chr1_+_161305558
|
0.989
|
NM_001113381
|
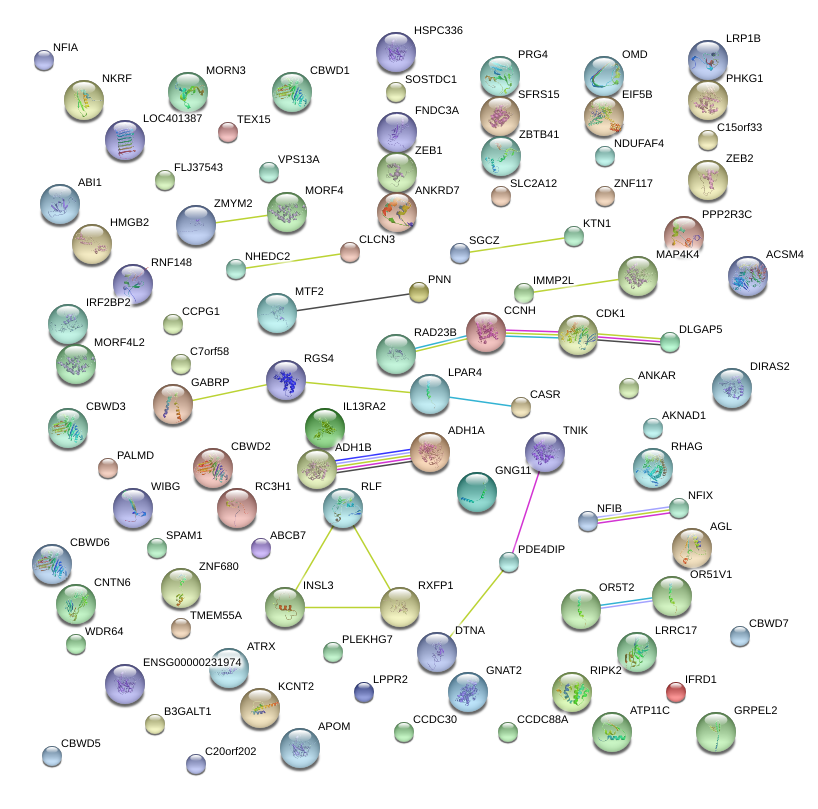
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_+_7348194
|
0.969
|
NM_001080454
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4
|
|
chr8_-_30826074
|
0.903
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr14_-_34660902
|
0.880
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr7_+_123353144
|
0.846
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr2_-_144994349
|
0.840
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_+_91654395
|
0.831
|
NM_001004330
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7
|
|
chr1_-_232810111
|
0.812
|
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr4_-_100431093
|
0.802
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr1_+_100089118
|
0.797
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chrX_-_114158388
|
0.783
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr9_-_94226380
|
0.781
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr1_-_194844061
|
0.767
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr7_-_122130201
|
0.761
|
NM_198085
|
RNF148
|
ring finger protein 148
|
|
chrX_-_153698930
|
0.709
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr7_-_63660889
|
0.705
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr21_-_31995972
|
0.689
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chrX_-_74292800
|
0.683
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chr1_+_40399618
|
0.664
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chrX_-_102828238
|
0.660
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr11_-_5178505
|
0.652
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr1_+_40399642
|
0.652
|
|
RLF
|
rearranged L-myc fusion
|
|
chr4_+_170817787
|
0.643
|
|
CLCN3
|
chloride channel 3
|
|
chr15_-_47700342
|
0.636
|
NM_152647
|
C15orf33
|
chromosome 15 open reading frame 33
|
|
chr15_-_53440069
|
0.634
|
|
CCPG1
|
cell cycle progression 1
|
|
chr2_+_168383427
|
0.631
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr20_+_1132097
|
0.629
|
NM_001009612
|
C20orf202
|
chromosome 20 open reading frame 202
|
|
chr14_+_38720085
|
0.629
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr3_+_1109619
|
0.623
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr4_-_174774368
|
0.607
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr14_+_55148488
|
0.606
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chrX_-_102829621
|
0.596
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chrX_-_76824726
|
0.594
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr9_+_79158164
|
0.589
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr1_+_99884018
|
0.585
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr7_-_63660766
|
0.584
|
|
ZNF680
|
zinc finger protein 680
|
|
chr5_+_148711419
|
0.580
|
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr18_-_35634247
|
0.573
|
|
LOC647946
|
hypothetical LOC647946
|
|
chr7_-_91632525
|
0.573
|
NM_001161528
|
LOC401387
|
leucine-rich repeat and death domain-containing protein
|
|
chr1_-_109201228
|
0.569
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chrX_-_102829595
|
0.560
|
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr5_-_86739683
|
0.558
|
|
CCNH
|
cyclin H
|
|
chr1_-_109957195
|
0.556
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr7_+_120415958
|
0.545
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr4_+_159662349
|
0.530
|
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr8_-_15140162
|
0.530
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr7_-_64088848
|
0.530
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr2_+_101874765
|
0.517
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr9_+_109131814
|
0.516
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr7_+_93388951
|
0.515
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr2_-_55413253
|
0.515
|
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr4_-_174491270
|
0.512
|
NM_001130689
|
HMGB2
|
high-mobility group box 2
|
|
chr7_+_120416103
|
0.512
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr9_-_14912419
|
0.504
|
|
|
|
|
chrX_+_56772391
|
0.502
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr1_-_195436258
|
0.502
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr7_-_110989582
|
0.500
|
NM_032549
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr6_-_49712483
|
0.499
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr6_-_68655708
|
0.499
|
|
|
|
|
chr1_+_19807168
|
0.497
|
|
|
|
|
chrX_-_118610880
|
0.496
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr14_-_54720101
|
0.496
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr11_-_55757236
|
0.492
|
NM_001004746
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2
|
|
chr2_+_190248955
|
0.487
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr11_+_65026382
|
0.486
|
|
|
|
|
chr2_-_142605739
|
0.486
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr2_+_99344060
|
0.485
|
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr5_+_170143289
|
0.485
|
NM_014211
|
GABRP
|
gamma-aminobutyric acid (GABA) A receptor, pi
|
|
chr2_+_101874809
|
0.483
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr12_-_54607925
|
0.482
|
|
WIBG
|
within bgcn homolog (Drosophila)
|
|
chr8_+_90839336
|
0.475
|
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chrX_-_138742050
|
0.474
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr1_+_93317374
|
0.474
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr14_+_38719462
|
0.470
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr10_+_62208094
|
0.464
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr1_+_61642287
|
0.463
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_172228673
|
0.462
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chrX_-_135783496
|
0.460
|
|
|
|
|
chr7_-_16471817
|
0.460
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chrX_+_77889861
|
0.460
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr7_+_117651947
|
0.460
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr1_+_184532027
|
0.458
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr9_+_70046658
|
0.456
|
NM_201453
|
CBWD5
CBWD3
CBWD1
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 1
COBW domain containing 6
|
|
chr5_+_60969392
|
0.456
|
NM_173667
|
FLJ37543
|
hypothetical protein FLJ37543
|
|
chr6_-_134415311
|
0.445
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr9_-_92444886
|
0.442
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr1_+_42773146
|
0.439
|
NM_001080850
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr7_+_102340579
|
0.437
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr6_-_97452481
|
0.436
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr1_-_143786993
|
0.435
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_+_239882202
|
0.433
|
NM_144625
|
WDR64
|
WD repeat domain 64
|
|
chr6_+_31731647
|
0.432
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr13_+_19430881
|
0.431
|
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr7_+_111879348
|
0.430
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr8_-_92122219
|
0.427
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr13_+_48582389
|
0.425
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr4_-_104217618
|
0.421
|
NM_178833
|
NHEDC2
|
Na+/H+ exchanger domain containing 2
|
|
chr18_+_30544193
|
0.421
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_-_127817143
|
0.418
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr1_-_110752068
|
0.416
|
NM_006402
|
HBXIP
|
hepatitis B virus x interacting protein
|
|
chrX_+_119622570
|
0.415
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_157822355
|
0.410
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr2_-_174538675
|
0.409
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr2_+_128644510
|
0.407
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr1_-_115070413
|
0.407
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr1_+_160002746
|
0.407
|
|
ATF6
|
activating transcription factor 6
|
|
chr14_+_51396799
|
0.406
|
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr1_+_15858950
|
0.404
|
NM_006511
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1
|
|
chr1_+_78888064
|
0.403
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr1_+_39569396
|
0.403
|
NM_033044
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr6_+_147568795
|
0.402
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_+_31731667
|
0.402
|
|
APOM
|
apolipoprotein M
|
|
chr2_+_114390975
|
0.400
|
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast)
|
|
chr1_-_47428310
|
0.400
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr14_+_51397134
|
0.399
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr5_+_109053507
|
0.398
|
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr4_-_164473107
|
0.398
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr12_+_49922592
|
0.394
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr4_+_144654526
|
0.393
|
|
|
|
|
chr4_-_80079393
|
0.390
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chrX_+_119622012
|
0.390
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_178100204
|
0.390
|
|
|
|
|
chr1_-_47428190
|
0.386
|
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr6_-_46933901
|
0.385
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chrX_+_123062143
|
0.384
|
|
STAG2
|
stromal antigen 2
|
|
chr6_+_88024152
|
0.384
|
|
ZNF292
|
zinc finger protein 292
|
|
chr11_-_5819702
|
0.382
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr3_+_99465777
|
0.382
|
NM_001005479
|
OR5H6
|
olfactory receptor, family 5, subfamily H, member 6
|
|
chr16_+_20682812
|
0.382
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr6_-_117096594
|
0.377
|
NM_145062
|
ZUFSP
|
zinc finger with UFM1-specific peptidase domain
|
|
chr1_+_151117421
|
0.377
|
NM_030663
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr11_+_109468734
|
0.376
|
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr6_+_142510094
|
0.375
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr6_-_135860479
|
0.375
|
|
AHI1
|
Abelson helper integration site 1
|
|
chr1_+_67923660
|
0.374
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr1_-_143075560
|
0.372
|
NM_178230
NM_001143883
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4B
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4B
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr8_+_39561243
|
0.372
|
NM_001190956
NM_014237
|
ADAM18
|
ADAM metallopeptidase domain 18
|
|
chr1_+_143807748
|
0.371
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr11_-_5419358
|
0.371
|
NM_001005288
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1
|
|
chr21_-_26867138
|
0.371
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr3_-_197293331
|
0.370
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr7_+_111850367
|
0.370
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr6_-_99955987
|
0.369
|
|
SFRS18
|
splicing factor, arginine/serine-rich 18
|
|
chr20_-_30703219
|
0.368
|
NM_182584
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr22_-_37211467
|
0.367
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr1_+_170768635
|
0.366
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr3_-_197293355
|
0.365
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr14_+_31615922
|
0.365
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr6_+_76367944
|
0.364
|
|
SENP6
|
SUMO1/sentrin specific peptidase 6
|
|
chr16_+_10430225
|
0.363
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr1_-_191422346
|
0.363
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr12_+_67366989
|
0.362
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr3_+_25445071
|
0.362
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr1_+_245779018
|
0.361
|
NM_145278
|
C1orf150
|
chromosome 1 open reading frame 150
|
|
chr4_-_153493133
|
0.361
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr3_-_197293394
|
0.357
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr10_+_111957292
|
0.355
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr14_+_95915774
|
0.354
|
NM_016472
|
C14orf129
|
chromosome 14 open reading frame 129
|
|
chr1_-_239865724
|
0.354
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr10_+_96433240
|
0.353
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr17_+_63338500
|
0.353
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr4_-_88531353
|
0.352
|
NM_016245
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr11_+_11820096
|
0.352
|
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr6_+_142510075
|
0.352
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr7_+_32501665
|
0.351
|
NM_015060
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr4_+_25931745
|
0.350
|
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr5_+_27508139
|
0.349
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr1_+_246432954
|
0.349
|
NM_001004689
|
OR2M3
|
olfactory receptor, family 2, subfamily M, member 3
|
|
chr5_-_43550891
|
0.348
|
NM_198566
|
C5orf34
|
chromosome 5 open reading frame 34
|
|
chr2_-_99412694
|
0.347
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr2_-_162808123
|
0.347
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr13_+_96592051
|
0.346
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chrX_-_102396704
|
0.345
|
NM_001006684
NM_153333
|
TCEAL8
|
transcription elongation factor A (SII)-like 8
|
|
chr11_+_65027721
|
0.343
|
|
|
|
|
chrX_+_119621933
|
0.342
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr4_+_90252990
|
0.341
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr15_-_75259714
|
0.341
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr6_-_76027286
|
0.340
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr11_+_27019084
|
0.340
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr2_+_48649662
|
0.340
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr4_-_88531245
|
0.338
|
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chrX_-_31194867
|
0.336
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chrX_-_132947141
|
0.335
|
|
GPC3
|
glypican 3
|
|
chr7_+_123272458
|
0.333
|
NM_012269
|
HYAL4
|
hyaluronoglucosaminidase 4
|
|
chr12_+_74160779
|
0.332
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr12_-_88573949
|
0.331
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr2_-_152538694
|
0.330
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr7_+_141124710
|
0.330
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chr2_+_122229590
|
0.330
|
NM_004622
|
TSN
|
translin
|
|
chr4_-_101658094
|
0.330
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr3_+_143858428
|
0.329
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr4_+_124537334
|
0.329
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_-_140280031
|
0.327
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr7_-_7724762
|
0.327
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr12_-_68379331
|
0.326
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr13_-_101852028
|
0.326
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr3_-_25658829
|
0.325
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr1_-_232810443
|
0.325
|
|
|
|
|
chr1_-_191341866
|
0.324
|
NM_016066
|
GLRX2
|
glutaredoxin 2
|
|
chr5_-_42847716
|
0.323
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|