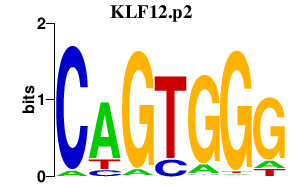
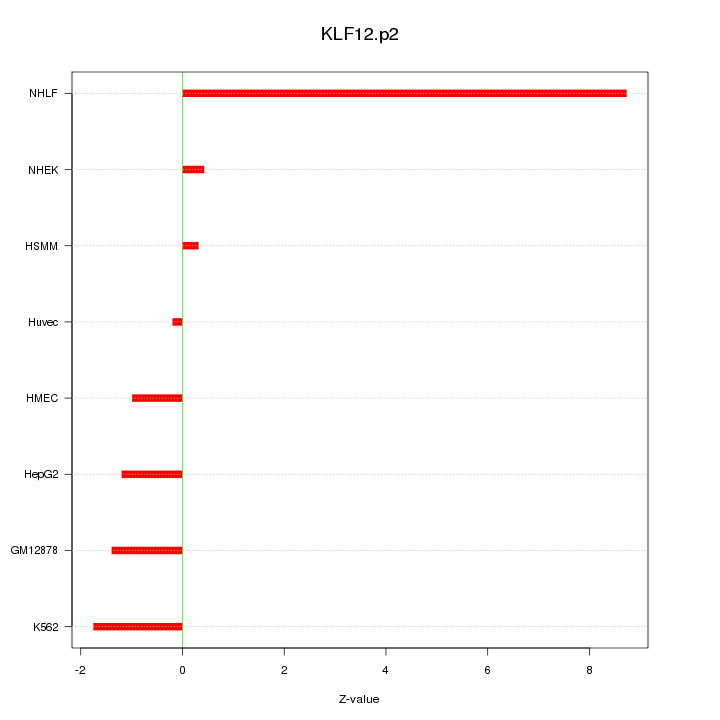
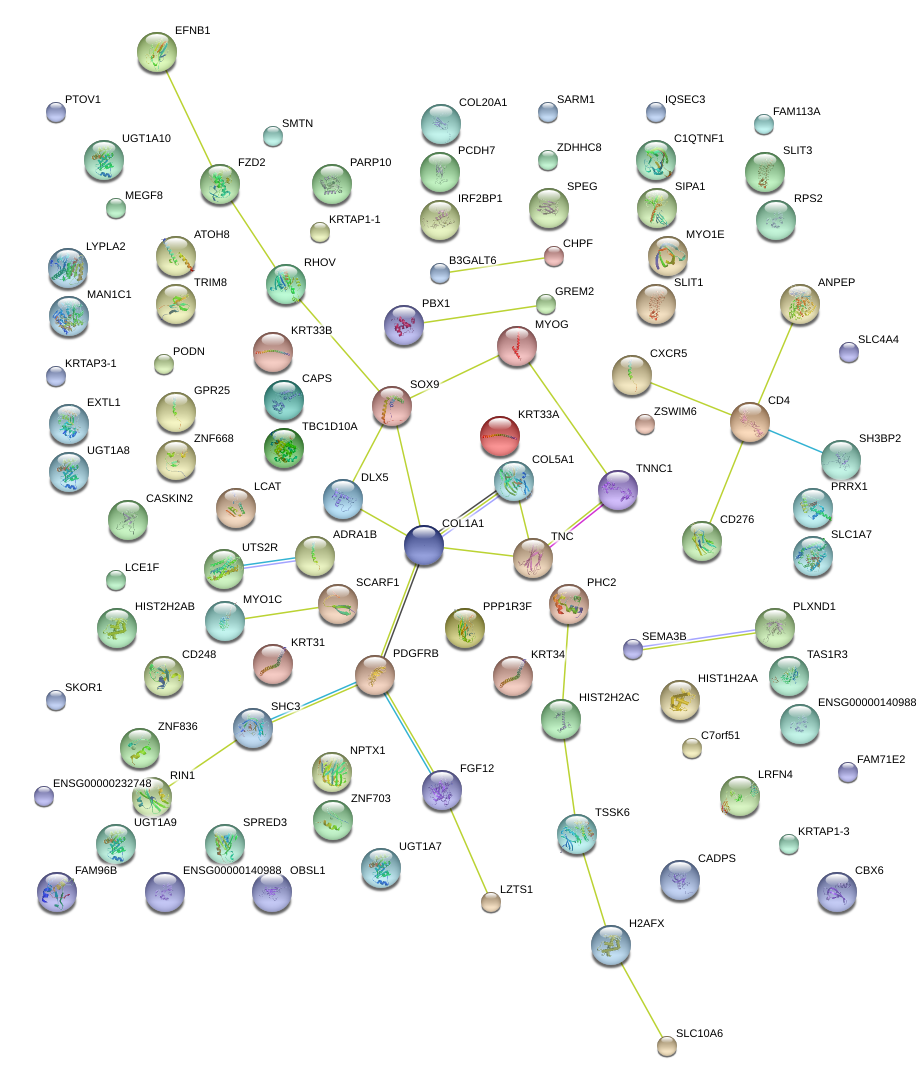
|
chr17_-_45620911
|
5.322
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr9_+_136673534
|
3.830
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr9_+_136673464
|
3.191
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr19_-_19487237
|
2.622
|
NM_032037
|
TSSK6
|
testis-specific serine kinase 6
|
|
chr1_-_53380709
|
2.560
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr16_-_30992574
|
2.346
|
|
ZNF668
|
zinc finger protein 668
|
|
chr2_-_220144254
|
2.322
|
|
OBSL1
|
obscurin-like 1
|
|
chr11_+_66381451
|
2.303
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr1_+_1256585
|
2.292
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr1_-_201321711
|
2.229
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr11_-_65841077
|
2.226
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr8_+_37672427
|
2.211
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr17_+_45617676
|
2.199
|
|
|
|
|
chr2_+_85834111
|
2.155
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr17_+_23723180
|
2.068
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr17_+_39990124
|
2.059
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr1_+_23990228
|
2.052
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr16_-_66535457
|
2.042
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr3_-_130761967
|
2.033
|
|
PLXND1
|
plexin D1
|
|
chr11_+_65164963
|
2.003
|
|
|
|
|
chrX_+_67965517
|
2.000
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr8_-_20156971
|
1.969
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr17_-_76064842
|
1.911
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr15_-_88148177
|
1.884
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr11_-_65860450
|
1.844
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr15_+_65904994
|
1.809
|
NM_001031807
|
SKOR1
|
SKI family transcriptional corepressor 1
|
|
chr19_+_43572679
|
1.778
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr20_-_2769120
|
1.763
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr1_+_168899936
|
1.744
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr1_+_162795538
|
1.742
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr17_-_1342700
|
1.734
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr17_-_36807322
|
1.699
|
NM_002277
|
KRT31
|
keratin 31
|
|
chr1_+_151015471
|
1.688
|
NM_178354
|
LCE1F
|
late cornified envelope 1F
|
|
chr20_+_61394982
|
1.660
|
NM_020882
|
COL20A1
|
collagen, type XX, alpha 1
|
|
chr20_-_2769245
|
1.651
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr4_+_2783764
|
1.649
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr5_-_168660293
|
1.615
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr5_+_133870175
|
1.604
|
|
|
|
|
chr9_-_116920232
|
1.580
|
NM_002160
|
TNC
|
tenascin C
|
|
chr2_+_234209838
|
1.564
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr4_+_72321113
|
1.560
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_-_149515331
|
1.550
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_+_46133
|
1.549
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr16_-_30993004
|
1.529
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr3_+_50281309
|
1.517
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr2_+_220017617
|
1.476
|
|
SPEG
|
SPEG complex locus
|
|
chr1_-_33613729
|
1.474
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr11_+_118259684
|
1.470
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr1_-_201321999
|
1.466
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr15_-_38953628
|
1.455
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr22_-_37598115
|
1.437
|
|
CBX6
|
chromobox homolog 6
|
|
chr2_-_220116508
|
1.431
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr5_+_60663842
|
1.413
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr17_-_36451189
|
1.410
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr11_-_118471313
|
1.401
|
|
H2AFX
|
H2A histone family, member X
|
|
chr19_-_60566419
|
1.400
|
NM_001145402
|
FAM71E2
|
family with sequence similarity 71, member E2
|
|
chr1_+_53300410
|
1.391
|
NM_153703
|
PODN
|
podocan
|
|
chr12_+_6768866
|
1.390
|
NM_000616
|
CD4
|
CD4 molecule
|
|
chr17_-_1495742
|
1.385
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr1_-_238842011
|
1.371
|
|
GREM2
|
gremlin 2
|
|
chr22_+_18499354
|
1.363
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr7_+_99919485
|
1.362
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr8_-_145132582
|
1.360
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr5_+_159276317
|
1.359
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr17_+_23722689
|
1.348
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr17_-_36418869
|
1.346
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr17_+_77925441
|
1.343
|
NM_018949
|
UTS2R
|
urotensin 2 receptor
|
|
chr22_-_29052863
|
1.330
|
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr1_+_199108705
|
1.323
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr11_+_65164767
|
1.319
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr9_-_90983501
|
1.316
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr17_+_67628749
|
1.314
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr19_-_51081147
|
1.301
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr19_+_55045754
|
1.297
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr17_+_74542066
|
1.296
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_+_25816545
|
1.280
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr3_-_193609531
|
1.280
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr22_+_29817218
|
1.270
|
|
SMTN
|
smoothelin
|
|
chr19_+_50963484
|
1.259
|
|
|
|
|
chr4_+_30331289
|
1.258
|
|
PCDH7
|
protocadherin 7
|
|
chr19_+_47521600
|
1.251
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr5_-_50714836
|
1.234
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr4_-_87989291
|
1.223
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr5_+_140762551
|
1.223
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr15_+_71763674
|
1.222
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|
|
chr11_-_118471385
|
1.221
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr1_+_26220895
|
1.221
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chrX_+_49013424
|
1.211
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr4_-_14613030
|
1.211
|
|
LOC441009
|
hypothetical protein LOC441009
|
|
chr10_+_104394185
|
1.203
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr19_+_5865192
|
1.200
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr16_-_1954827
|
1.195
|
NM_002952
|
RPS2
|
ribosomal protein S2
|
|
chr7_-_96492064
|
1.188
|
NM_005221
|
DLX5
|
distal-less homeobox 5
|
|
chr1_+_1157485
|
1.180
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chr12_-_49878216
|
1.175
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr20_+_31268779
|
1.172
|
NM_001042439
NM_178466
|
C20orf71
|
chromosome 20 open reading frame 71
|
|
chr9_-_90983195
|
1.171
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr15_+_75074483
|
1.170
|
NM_003978
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr10_+_88404285
|
1.168
|
NM_001030015
NM_033282
|
OPN4
|
opsin 4
|
|
chr22_+_29817315
|
1.166
|
|
SMTN
|
smoothelin
|
|
chr15_+_72005864
|
1.164
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr5_+_140790141
|
1.163
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr19_-_50963942
|
1.155
|
|
|
|
|
chr9_-_113130533
|
1.151
|
NM_205859
|
OR2K2
|
olfactory receptor, family 2, subfamily K, member 2
|
|
chr20_+_2769338
|
1.149
|
NM_022575
NM_080413
|
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae)
|
|
chr17_-_35331435
|
1.148
|
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr2_+_219453487
|
1.143
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr15_-_63147228
|
1.136
|
|
RASL12
|
RAS-like, family 12
|
|
chr17_-_38081967
|
1.135
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr19_+_60679510
|
1.125
|
NM_033113
|
ZNF628
|
zinc finger protein 628
|
|
chr22_-_37598160
|
1.118
|
|
CBX6
|
chromobox homolog 6
|
|
chr7_+_27190688
|
1.110
|
|
|
|
|
chr6_-_34324739
|
1.109
|
|
C6orf1
|
chromosome 6 open reading frame 1
|
|
chr10_-_105604942
|
1.108
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr5_+_140790510
|
1.103
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr15_-_72446979
|
1.103
|
NM_000781
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr19_+_2427122
|
1.101
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr11_-_1464386
|
1.099
|
NM_001172223
|
MOB2
|
Mps one binder kinase activator-like 2
|
|
chr9_+_134447795
|
1.095
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr5_+_140790310
|
1.091
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr6_-_152744022
|
1.091
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr5_+_140698537
|
1.088
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr12_-_51861388
|
1.087
|
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr1_-_68470585
|
1.087
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr15_-_63147350
|
1.085
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr17_+_7549138
|
1.084
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr19_-_55072263
|
1.081
|
NM_032375
NM_001098633
NM_001098632
|
AKT1S1
|
AKT1 substrate 1 (proline-rich)
|
|
chr7_+_27248688
|
1.071
|
NM_001989
|
EVX1
|
even-skipped homeobox 1
|
|
chr16_-_30992323
|
1.070
|
NM_001172668
|
ZNF668
|
zinc finger protein 668
|
|
chr15_+_72005875
|
1.067
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr10_-_105410840
|
1.060
|
|
|
|
|
chr9_+_110664366
|
1.056
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chr16_-_2099854
|
1.056
|
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr10_-_81698840
|
1.054
|
NM_003019
|
SFTPD
|
surfactant protein D
|
|
chr20_-_604822
|
1.054
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr19_+_16296734
|
1.047
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr2_+_220015016
|
1.041
|
|
SPEG
|
SPEG complex locus
|
|
chr11_+_46323495
|
1.040
|
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr20_+_34603311
|
1.040
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr20_-_2769789
|
1.037
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chrX_-_47364173
|
1.036
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr11_+_5925152
|
1.034
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr12_-_79855618
|
1.031
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr7_+_75947762
|
1.029
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr7_+_100111712
|
1.028
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr2_-_228754585
|
1.023
|
NM_001142644
NM_030623
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing
|
|
chr17_-_17816425
|
1.023
|
NM_001033551
NM_001082968
|
TOM1L2
|
target of myb1-like 2 (chicken)
|
|
chr20_-_62181588
|
1.020
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr12_+_6930778
|
1.015
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr19_+_2173415
|
1.014
|
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr1_-_238842071
|
1.011
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr22_+_35777561
|
1.007
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr3_+_42670179
|
1.006
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr9_+_115970599
|
1.006
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr4_+_7245218
|
1.006
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr17_+_23723220
|
1.005
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr5_+_140790365
|
1.002
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr7_-_5427367
|
1.001
|
|
|
|
|
chr10_+_124310170
|
0.992
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr11_+_6847499
|
0.991
|
NM_001004460
|
OR10A2
|
olfactory receptor, family 10, subfamily A, member 2
|
|
chr2_+_241279934
|
0.988
|
NM_198998
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr16_+_2503915
|
0.984
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr9_+_35528890
|
0.981
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr16_+_30583599
|
0.972
|
|
FBRS
|
fibrosin
|
|
chr11_-_1599900
|
0.969
|
NM_001012709
|
KRTAP5-4
|
keratin associated protein 5-4
|
|
chr7_-_117300763
|
0.966
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr5_+_140709907
|
0.964
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr1_-_36562341
|
0.963
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chrX_-_53367246
|
0.961
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr9_+_115303527
|
0.960
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr12_+_52653176
|
0.960
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr21_-_46428788
|
0.958
|
NM_001142854
NM_032261
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr20_+_19818209
|
0.958
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chrX_-_106033202
|
0.955
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr3_+_50281582
|
0.953
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr2_-_128138435
|
0.952
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr16_-_65510230
|
0.952
|
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr20_+_42007949
|
0.951
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr17_-_34607257
|
0.946
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr19_-_14447122
|
0.945
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr3_+_49033621
|
0.945
|
NM_199073
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr10_+_99463446
|
0.943
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr9_+_138367329
|
0.942
|
NM_001145639
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr9_-_138387926
|
0.939
|
NM_052813
NM_052814
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr4_+_30330963
|
0.939
|
NM_001173523
NM_002589
NM_032456
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr17_+_23723113
|
0.933
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr2_-_220144511
|
0.930
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr12_-_92489652
|
0.928
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr5_+_149717489
|
0.927
|
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr8_+_8597075
|
0.927
|
NM_194284
|
CLDN23
|
claudin 23
|
|
chr16_+_30844707
|
0.924
|
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr2_-_73373956
|
0.920
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr12_+_7038240
|
0.919
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr2_+_238673689
|
0.916
|
NM_194312
|
ESPNL
|
espin-like
|
|
chrX_-_152889815
|
0.913
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr16_-_66535923
|
0.909
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr6_-_110843432
|
0.909
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr9_-_139128979
|
0.908
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr15_-_87257553
|
0.907
|
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr19_-_3928516
|
0.906
|
|
EEF2
|
eukaryotic translation elongation factor 2
|