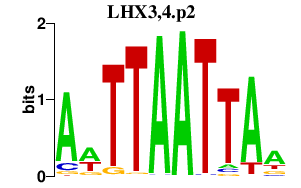
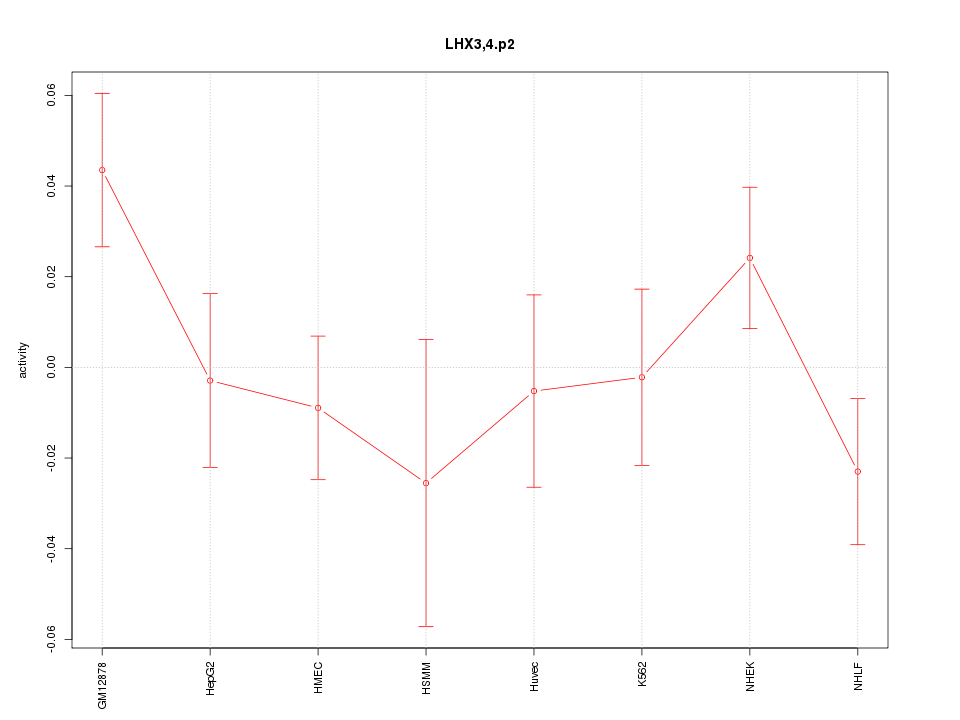
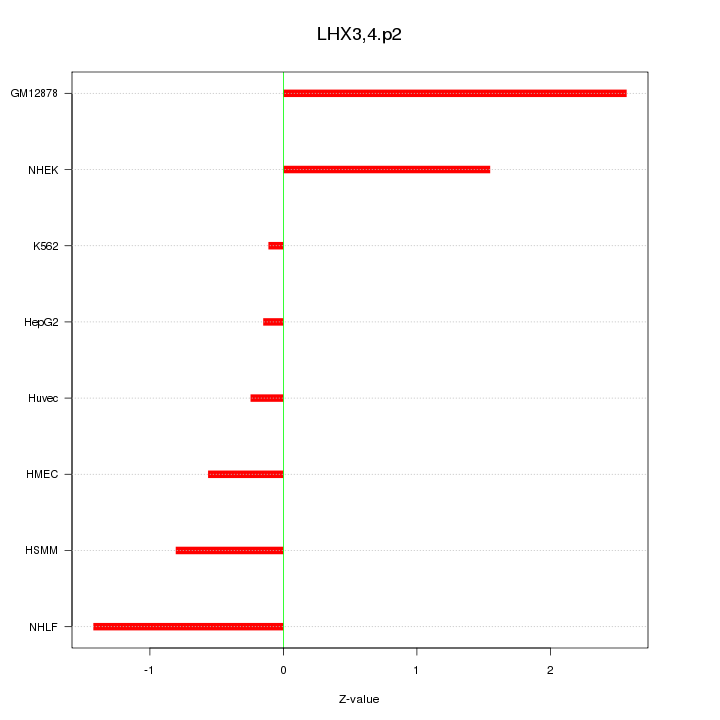
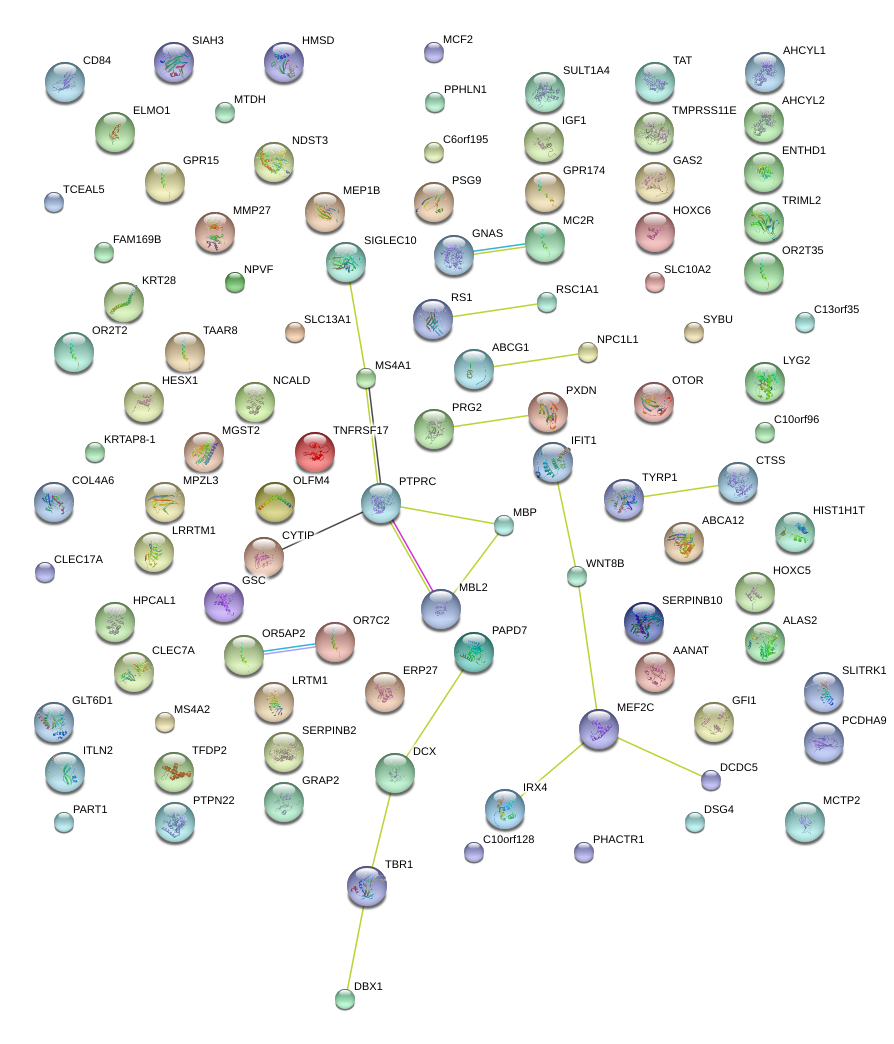
|
chr3_-_57209319
|
1.696
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr13_-_83354425
|
1.164
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr13_+_112349358
|
1.156
|
NM_207440
|
C13orf35
|
chromosome 13 open reading frame 35
|
|
chr9_-_137671206
|
1.120
|
NM_182974
|
GLT6D1
|
glycosyltransferase 6 domain containing 1
|
|
chr7_-_25234582
|
1.048
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr22_+_38627031
|
1.007
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr19_-_56612653
|
0.957
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr11_-_30910074
|
0.940
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr4_+_68995761
|
0.887
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr4_-_189263395
|
0.859
|
NM_173553
|
TRIML2
|
tripartite motif family-like 2
|
|
chr7_-_36991189
|
0.851
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_+_119174947
|
0.836
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr5_-_1935764
|
0.827
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr18_-_13905534
|
0.811
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr1_+_196874839
|
0.810
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_11966464
|
0.795
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr18_+_59767567
|
0.779
|
NM_001123366
|
HMSD
|
histocompatibility (minor) serpin domain containing
|
|
chr2_-_99238001
|
0.718
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr6_+_132915522
|
0.716
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr11_-_102081654
|
0.713
|
NM_022122
|
MMP27
|
matrix metallopeptidase 27
|
|
chr18_+_28023984
|
0.708
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr13_+_52500830
|
0.706
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr10_+_91142301
|
0.700
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr11_+_22646229
|
0.695
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr19_+_14554895
|
0.686
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr2_-_215711313
|
0.676
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr7_-_44547382
|
0.660
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr15_+_92700167
|
0.654
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr20_+_56919233
|
0.645
|
|
GNAS
|
GNAS complex locus
|
|
chr11_-_117628211
|
0.645
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr1_-_92725017
|
0.642
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr13_-_45323757
|
0.636
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr22_-_38619739
|
0.627
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr14_-_94306104
|
0.621
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr15_-_96875133
|
0.611
|
NM_182562
|
FAM169B
|
family with sequence similarity 169, member B
|
|
chr18_-_72857665
|
0.607
|
|
MBP
|
myelin basic protein
|
|
chr2_+_161980865
|
0.587
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr3_-_54937001
|
0.579
|
NM_020678
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1
|
|
chr6_+_12825818
|
0.571
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr18_+_59733724
|
0.567
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr7_-_122627235
|
0.567
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr2_+_161981046
|
0.558
|
|
TBR1
|
T-box, brain, 1
|
|
chr8_+_98772537
|
0.557
|
|
MTDH
|
metadherin
|
|
chrX_-_55074125
|
0.546
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr1_-_158815895
|
0.536
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr3_+_99733432
|
0.531
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr13_-_83354474
|
0.529
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chrX_-_138552517
|
0.526
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr1_-_114215768
|
0.523
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chrX_-_110542040
|
0.520
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr20_+_16676997
|
0.516
|
NM_020157
|
OTOR
|
otoraplin
|
|
chrX_+_78313124
|
0.513
|
NM_032553
|
GPR174
|
G protein-coupled receptor 174
|
|
chr10_+_118073929
|
0.513
|
NM_198515
|
C10orf96
|
chromosome 10 open reading frame 96
|
|
chr2_-_80385227
|
0.512
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr10_+_102212787
|
0.503
|
NM_003393
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr22_+_38627129
|
0.501
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr12_-_10173947
|
0.496
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr11_+_59979857
|
0.491
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr15_-_64108718
|
0.490
|
|
|
|
|
chr6_-_25679197
|
0.487
|
|
|
|
|
chrX_-_18600108
|
0.482
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr16_-_70168449
|
0.480
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr4_+_140806536
|
0.480
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr8_-_102872354
|
0.478
|
|
NCALD
|
neurocalcin delta
|
|
chrX_-_102418329
|
0.476
|
NM_001012979
|
TCEAL5
|
transcription elongation factor A (SII)-like 5
|
|
chr17_-_36209672
|
0.475
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr1_-_159191184
|
0.473
|
NM_080878
|
ITLN2
|
intelectin 2
|
|
chr13_-_102517196
|
0.469
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr6_-_26216299
|
0.469
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr1_-_149004916
|
0.468
|
NM_004079
|
CTSS
|
cathepsin S
|
|
chr2_-_158008763
|
0.467
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr8_-_110762409
|
0.464
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_-_20138445
|
0.461
|
NM_001029865
|
DBX1
|
developing brain homeobox 1
|
|
chr10_-_50066386
|
0.455
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr18_+_27210737
|
0.455
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr1_+_196874867
|
0.452
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_101398429
|
0.447
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr21_+_42492867
|
0.445
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr7_-_37454933
|
0.444
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr11_-_56166490
|
0.441
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr1_+_246682699
|
0.437
|
NM_001004136
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2
|
|
chr21_-_31107431
|
0.437
|
NM_175857
|
KRTAP8-1
|
keratin associated protein 8-1
|
|
chr17_+_71975224
|
0.436
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr6_-_2580296
|
0.434
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr7_+_128802719
|
0.433
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr5_+_59819761
|
0.432
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr3_-_143230124
|
0.432
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr1_-_149004852
|
0.423
|
|
CTSS
|
cathepsin S
|
|
chr12_+_52697160
|
0.422
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr6_+_31538934
|
0.422
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chrX_-_107569256
|
0.418
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr12_-_14982708
|
0.412
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr9_+_12683385
|
0.411
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr5_+_140207540
|
0.410
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr5_-_88155360
|
0.410
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr14_-_46882144
|
0.409
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chrX_-_131089552
|
0.408
|
NM_194277
|
FRMD7
|
FERM domain containing 7
|
|
chr13_-_34948758
|
0.407
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chrX_+_37524250
|
0.404
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr2_+_158559936
|
0.401
|
NM_001135098
|
UPP2
|
uridine phosphorylase 2
|
|
chrX_+_50044279
|
0.401
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chrX_+_37524190
|
0.400
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr11_-_74478405
|
0.400
|
NM_001005285
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4
|
|
chr4_-_68511738
|
0.400
|
NM_001114387
NM_182606
|
TMPRSS11A
|
transmembrane protease, serine 11A
|
|
chr15_+_100175888
|
0.400
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr10_+_57028755
|
0.397
|
NM_001190478
|
MTRNR2L5
|
MT-RNR2-like 5
|
|
chr5_-_160211623
|
0.395
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr7_-_38279757
|
0.391
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr11_+_61794205
|
0.390
|
NM_002411
|
SCGB2A2
|
secretoglobin, family 2A, member 2
|
|
chr9_-_35962454
|
0.386
|
|
YBX1
|
Y box binding protein 1
|
|
chr11_-_89181390
|
0.383
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr4_+_88748704
|
0.380
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr1_-_151587645
|
0.380
|
NM_020393
|
PGLYRP4
|
peptidoglycan recognition protein 4
|
|
chr12_+_91620749
|
0.378
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr8_-_59575249
|
0.378
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr8_-_102872372
|
0.373
|
|
NCALD
|
neurocalcin delta
|
|
chr12_+_130004404
|
0.372
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr17_-_36347197
|
0.370
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_-_245942679
|
0.370
|
NM_001005286
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1
|
|
chr6_+_31604717
|
0.367
|
NM_001011700
|
MCCD1
|
mitochondrial coiled-coil domain 1
|
|
chr5_-_156705250
|
0.366
|
NM_001001343
|
FNDC9
|
fibronectin type III domain containing 9
|
|
chr3_+_82117978
|
0.366
|
|
|
|
|
chrX_+_48978875
|
0.364
|
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr21_-_31175722
|
0.364
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr20_+_5935897
|
0.361
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr6_-_50045296
|
0.361
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chr1_-_203592538
|
0.360
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr22_+_35290271
|
0.355
|
|
|
|
|
chr9_+_92603832
|
0.354
|
NM_001135052
NM_003177
|
SYK
|
spleen tyrosine kinase
|
|
chr8_-_110730183
|
0.354
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_-_101396459
|
0.353
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_9863929
|
0.353
|
|
GAS7
|
growth arrest-specific 7
|
|
chr8_+_124032830
|
0.353
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr6_+_167504070
|
0.352
|
NM_001145121
|
TCP10L2
TCP10
|
t-complex 10-like 2 (mouse)
t-complex 10 homolog (mouse)
|
|
chr10_-_21825879
|
0.348
|
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr12_+_52696908
|
0.345
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr14_-_106084519
|
0.345
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_-_79811475
|
0.341
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr3_-_152517204
|
0.341
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr10_-_48426868
|
0.340
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr1_-_149004879
|
0.340
|
|
CTSS
|
cathepsin S
|
|
chr15_+_32048380
|
0.339
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr5_-_22889192
|
0.339
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr11_+_89032112
|
0.339
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr2_+_161981138
|
0.338
|
|
TBR1
|
T-box, brain, 1
|
|
chr19_-_40995606
|
0.337
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chrX_+_85404626
|
0.335
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr12_+_52634950
|
0.332
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr20_-_7869068
|
0.330
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr1_+_173111306
|
0.329
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr18_+_27281729
|
0.329
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr8_+_92330691
|
0.328
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr6_-_136829705
|
0.327
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr17_-_9635325
|
0.327
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr8_-_7710547
|
0.327
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr15_+_70477707
|
0.326
|
NM_001080462
|
TMEM202
|
transmembrane protein 202
|
|
chr5_+_156628929
|
0.326
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_+_190394214
|
0.323
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr21_-_30460841
|
0.322
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr6_-_167717907
|
0.317
|
NM_004610
|
TCP10
|
t-complex 10 homolog (mouse)
|
|
chr12_-_16321885
|
0.315
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr4_+_74497998
|
0.315
|
|
ALB
|
albumin
|
|
chrX_+_115215985
|
0.311
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr1_-_157014048
|
0.311
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chrX_+_107174855
|
0.309
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr2_-_207291364
|
0.308
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr11_-_123182266
|
0.305
|
NM_001005325
|
OR6M1
|
olfactory receptor, family 6, subfamily M, member 1
|
|
chr8_-_7200278
|
0.304
|
|
FAM66B
|
family with sequence similarity 66, member B
|
|
chrX_-_24600661
|
0.302
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chrX_-_142432884
|
0.301
|
NM_001009609
|
SPANXN3
|
SPANX family, member N3
|
|
chr10_+_90511142
|
0.301
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chrX_+_140810117
|
0.300
|
NM_177456
|
MAGEC3
|
melanoma antigen family C, 3
|
|
chr6_-_33016561
|
0.298
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr7_+_147918589
|
0.298
|
NM_145304
|
C7orf33
|
chromosome 7 open reading frame 33
|
|
chr1_+_117345894
|
0.298
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr2_-_134042500
|
0.296
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chrX_-_13744933
|
0.296
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_+_134803450
|
0.293
|
NM_018666
|
SAGE1
|
sarcoma antigen 1
|
|
chrX_+_28515479
|
0.293
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr6_+_12825022
|
0.289
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr14_+_61532293
|
0.288
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr6_+_12824873
|
0.287
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr15_-_70838703
|
0.287
|
|
|
|
|
chr7_+_37689903
|
0.286
|
|
|
|
|
chr6_+_55300225
|
0.286
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr2_+_234490781
|
0.284
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr1_+_196874759
|
0.282
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_151549790
|
0.282
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr6_-_29635680
|
0.280
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr14_-_105997732
|
0.280
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_+_94097513
|
0.279
|
NM_006215
|
SERPINA4
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4
|
|
chr20_+_31334601
|
0.278
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr4_+_74498042
|
0.278
|
|
ALB
|
albumin
|
|
chr9_+_129066576
|
0.277
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr2_+_73864823
|
0.274
|
NM_001080474
|
C2orf78
|
chromosome 2 open reading frame 78
|
|
chrX_-_138875342
|
0.274
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr4_-_47678187
|
0.273
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr1_+_111634996
|
0.272
|
NM_021797
NM_201653
|
CHIA
|
chitinase, acidic
|
|
chr2_+_95621492
|
0.272
|
NM_138800
|
TRIM43
|
tripartite motif containing 43
|
|
chr10_-_21826192
|
0.271
|
|
C10orf114
|
chromosome 10 open reading frame 114
|