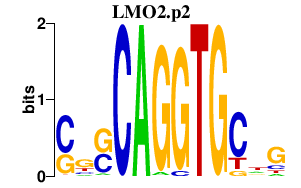
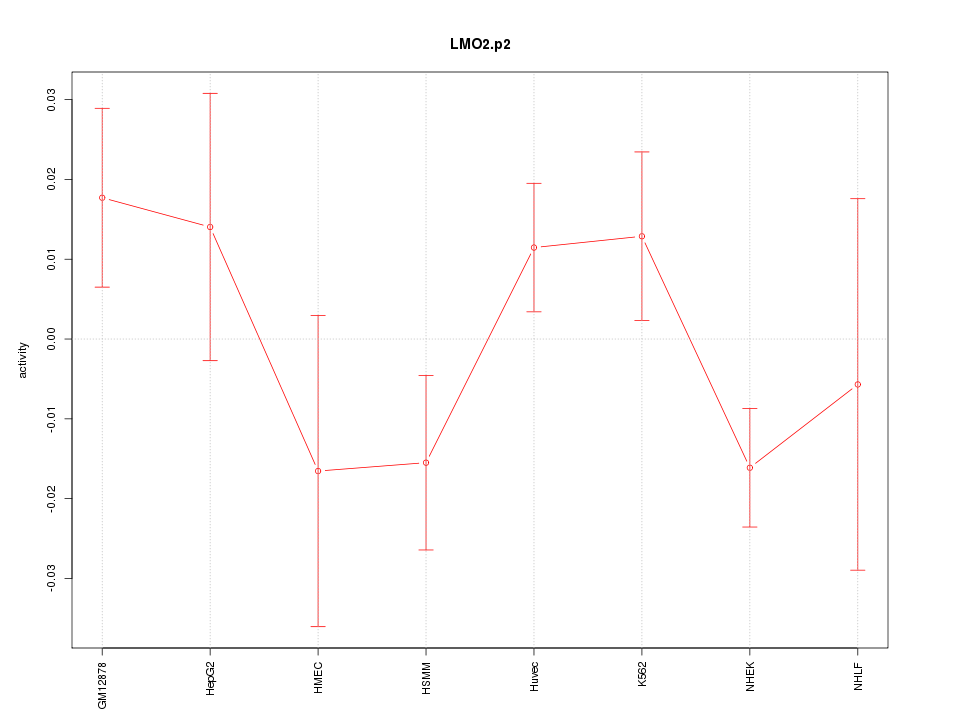
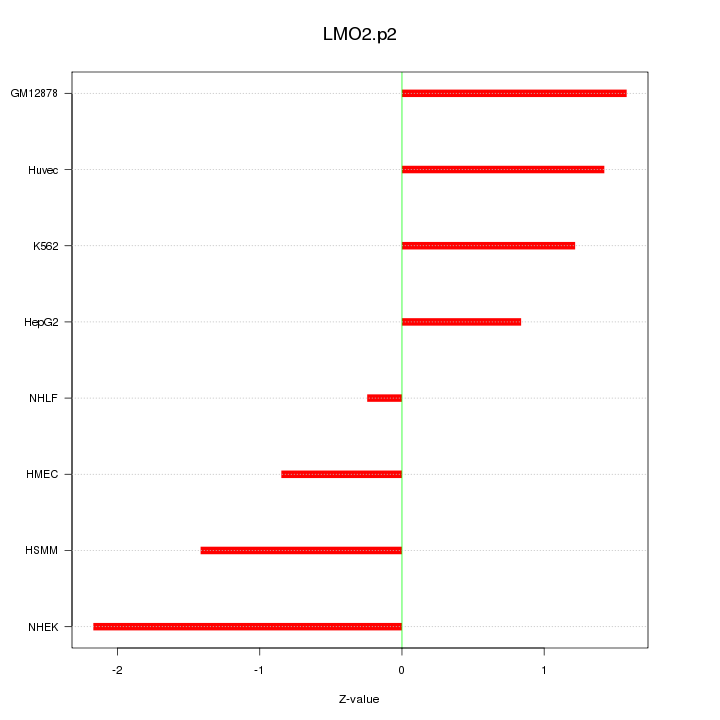
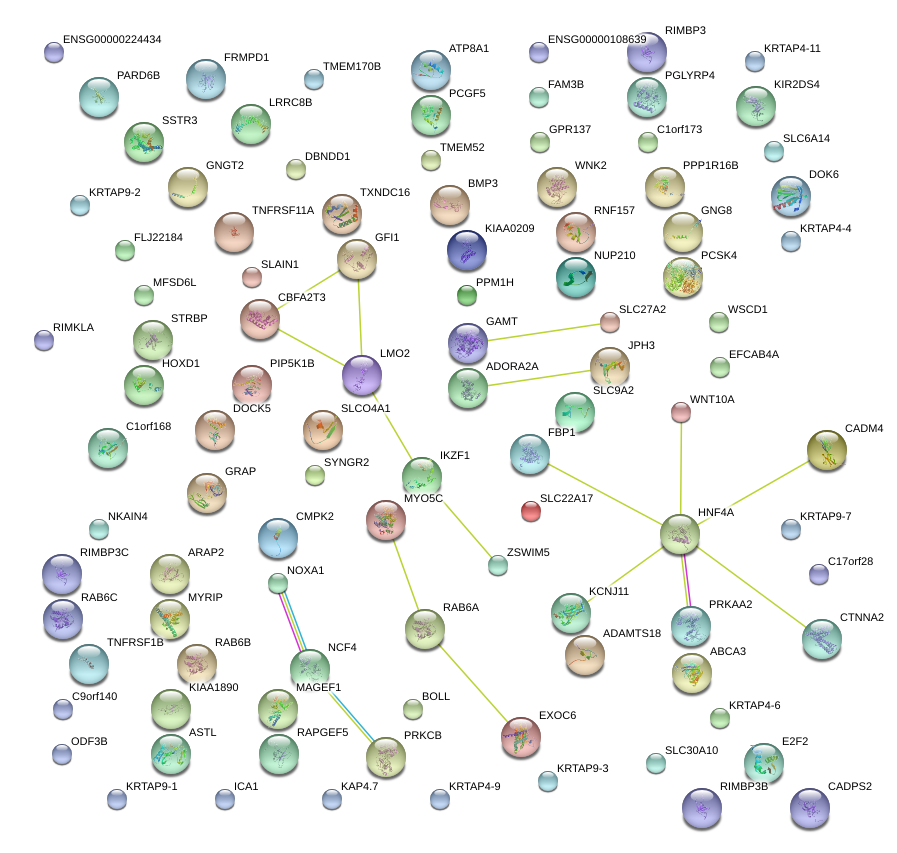
|
chr2_-_6923215
|
3.550
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr1_-_1840564
|
2.739
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr17_-_8643163
|
2.079
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr19_-_7845325
|
1.994
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr16_+_23754772
|
1.797
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr9_-_96441654
|
1.744
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr17_-_36570508
|
1.655
|
NM_032524
|
KRTAP4-4
|
keratin associated protein 4-4
|
|
chr17_-_36528113
|
1.606
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr13_+_77170470
|
1.592
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr2_+_79593679
|
1.583
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr9_+_37640996
|
1.572
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr3_+_39826032
|
1.533
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr7_-_8268292
|
1.435
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr17_+_36599664
|
1.426
|
NM_001190460
|
KRTAP9-1
|
keratin associated protein 9-1
|
|
chr4_-_42353640
|
1.408
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr17_+_36636404
|
1.406
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr2_+_79593567
|
1.394
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr16_+_23754860
|
1.366
|
|
PRKCB
|
protein kinase C, beta
|
|
chr16_-_76026431
|
1.357
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr17_+_36493927
|
1.327
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr3_-_13436802
|
1.318
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr11_-_33847507
|
1.301
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr16_+_23754822
|
1.295
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_-_74912009
|
1.292
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr20_+_48781457
|
1.285
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr20_-_61356247
|
1.284
|
NM_152864
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4
|
|
chr16_-_87570716
|
1.244
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr5_+_168996852
|
1.193
|
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr1_+_89762984
|
1.183
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr5_+_168996816
|
1.178
|
NM_004946
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr16_+_23754997
|
1.176
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_-_218168137
|
1.168
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr9_-_96441567
|
1.152
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr1_+_12149637
|
1.150
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr1_-_218168404
|
1.148
|
|
|
|
|
chr1_+_12149662
|
1.145
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr10_+_92970302
|
1.137
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr9_-_125070613
|
1.116
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr17_-_71747867
|
1.097
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr7_-_8268206
|
1.096
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr18_+_65219114
|
1.078
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr7_-_8268666
|
1.047
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr10_+_94598204
|
1.045
|
NM_019053
|
EXOC6
|
exocyst complex component 6
|
|
chr15_-_50375143
|
1.032
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr1_+_89763030
|
1.017
|
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr17_+_73676219
|
1.006
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr1_+_56883333
|
1.005
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr2_+_219453487
|
0.997
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr8_-_144862266
|
0.992
|
NM_001162914
|
LOC100130274
|
coiled-coil domain containing 121-like
|
|
chr20_+_42463279
|
0.961
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr9_+_70509925
|
0.958
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr22_+_23153529
|
0.953
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr9_-_125070675
|
0.951
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr2_-_96167900
|
0.945
|
NM_001002036
|
ASTL
|
astacin-like metallo-endopeptidase (M12 family)
|
|
chr3_-_185912240
|
0.939
|
NM_022149
|
MAGEF1
|
melanoma antigen family F, 1
|
|
chr4_+_82171142
|
0.934
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr14_-_22891263
|
0.934
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr1_-_218168548
|
0.928
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr7_-_22363036
|
0.896
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr17_+_5913149
|
0.879
|
|
WSCD1
|
WSC domain containing 1
|
|
chr16_-_2330738
|
0.875
|
NM_001089
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr20_+_36867708
|
0.872
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr2_+_170298487
|
0.867
|
|
|
|
|
chr16_-_88613287
|
0.865
|
NM_001042610
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr19_-_48835783
|
0.860
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr1_-_151587645
|
0.860
|
NM_020393
|
PGLYRP4
|
peptidoglycan recognition protein 4
|
|
chr7_+_50314801
|
0.859
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr9_+_139437624
|
0.858
|
NM_006647
|
NOXA1
|
NADPH oxidase activator 1
|
|
chr3_-_13436725
|
0.856
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr15_+_48261649
|
0.853
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr19_-_51829778
|
0.852
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr17_-_18890963
|
0.851
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr12_-_61615124
|
0.849
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_-_70480385
|
0.849
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr19_-_1441403
|
0.846
|
NM_017573
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chr1_-_92724181
|
0.836
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr9_-_139084833
|
0.835
|
NM_178448
|
C9orf140
|
chromosome 9 open reading frame 140
|
|
chr2_+_176761552
|
0.827
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr6_+_11646472
|
0.825
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_-_57057956
|
0.825
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chr12_-_61614930
|
0.810
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr1_-_45444812
|
0.803
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chrX_+_115481774
|
0.802
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr16_+_86193993
|
0.799
|
NM_020655
|
JPH3
|
junctophilin 3
|
|
chr3_-_199291938
|
0.797
|
|
LOC348840
|
hypothetical LOC348840
|
|
chr18_+_58143499
|
0.795
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr19_-_1441340
|
0.793
|
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chr4_-_35922288
|
0.789
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr22_+_35586950
|
0.787
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr9_+_94987032
|
0.785
|
NM_006648
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chr2_-_198359182
|
0.779
|
NM_197970
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr3_-_135096986
|
0.776
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr22_-_35938298
|
0.776
|
NM_001051
|
SSTR3
|
somatostatin receptor 3
|
|
chr22_-_49317408
|
0.770
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr21_+_41610530
|
0.769
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr11_+_817584
|
0.766
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr1_+_42619014
|
0.763
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr11_-_17366781
|
0.760
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr16_-_2330594
|
0.756
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr20_+_60744231
|
0.753
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr1_-_23729871
|
0.753
|
|
E2F2
|
E2F transcription factor 2
|
|
chr19_-_1352474
|
0.746
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr2_+_102602579
|
0.730
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr22_+_20068039
|
0.728
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr1_+_1062137
|
0.723
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr7_-_122313646
|
0.722
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr4_-_144841277
|
0.720
|
NM_001168235
|
FREM3
|
FRAS1 related extracellular matrix 3
|
|
chr17_-_8809656
|
0.717
|
NM_001142633
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr2_+_208979800
|
0.715
|
NM_005048
|
PTH2R
|
parathyroid hormone 2 receptor
|
|
chr7_+_12576727
|
0.715
|
NM_001112706
|
SCIN
|
scinderin
|
|
chr14_+_30413393
|
0.714
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr11_-_17367439
|
0.708
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr11_+_44026106
|
0.707
|
NM_001031854
|
ACCSL
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like
|
|
chr19_+_18145631
|
0.703
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr2_+_112612429
|
0.702
|
NM_001128165
NM_153214
|
FBLN7
|
fibulin 7
|
|
chrX_-_118711088
|
0.702
|
|
SEPT6
|
septin 6
|
|
chr2_+_242679507
|
0.702
|
|
LOC728323
|
hypothetical LOC728323
|
|
chr20_-_23014828
|
0.698
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr15_-_53822468
|
0.697
|
NM_173814
|
PRTG
|
protogenin
|
|
chr20_+_60744268
|
0.695
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr4_-_647424
|
0.693
|
|
|
|
|
chr17_+_78286687
|
0.690
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr11_-_64641700
|
0.686
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr17_-_20739924
|
0.685
|
NM_001004306
|
CCDC144NL
|
coiled-coil domain containing 144 family, N-terminal like
|
|
chr19_+_18145619
|
0.685
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr9_+_73954174
|
0.684
|
|
GDA
|
guanine deaminase
|
|
chr17_+_44564820
|
0.680
|
NM_001159388
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2
|
|
chr15_-_93671314
|
0.679
|
|
LOC400456
|
hypothetical LOC400456
|
|
chr2_+_198377561
|
0.677
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr10_-_98470164
|
0.675
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr1_-_85439315
|
0.673
|
NM_032184
|
SYDE2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans)
|
|
chr19_-_6643953
|
0.671
|
|
C3
|
complement component 3
|
|
chr7_-_960534
|
0.668
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr11_-_68505030
|
0.665
|
NM_198923
|
MRGPRD
|
MAS-related GPR, member D
|
|
chr1_+_65492679
|
0.664
|
|
LOC645195
DNAJC6
|
hypothetical LOC645195
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chrX_-_132947290
|
0.662
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr11_-_33847834
|
0.661
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr10_+_70881221
|
0.657
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr9_-_14683459
|
0.650
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr20_+_42463350
|
0.641
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_+_31647854
|
0.637
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr8_-_27906287
|
0.637
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr19_+_1033816
|
0.633
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr9_-_103397023
|
0.632
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr9_-_91302707
|
0.629
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr8_+_121206478
|
0.628
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr7_+_140420500
|
0.627
|
NM_001195278
|
LOC100131199
|
transmembrane protein 178-like
|
|
chr3_+_168936125
|
0.622
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr22_-_20314274
|
0.621
|
NM_001017964
|
YDJC
|
YdjC homolog (bacterial)
|
|
chr16_+_87232501
|
0.618
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr10_-_98935570
|
0.618
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr11_-_124137260
|
0.613
|
NM_138961
|
ESAM
|
endothelial cell adhesion molecule
|
|
chr7_+_45163891
|
0.612
|
NM_005856
|
RAMP3
|
receptor (G protein-coupled) activity modifying protein 3
|
|
chr14_-_22891865
|
0.611
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr16_-_2330712
|
0.608
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr1_+_95355415
|
0.606
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr16_+_76690664
|
0.603
|
NM_016373
NM_130791
NM_130844
|
WWOX
|
WW domain containing oxidoreductase
|
|
chr14_-_99695684
|
0.601
|
NM_206918
|
DEGS2
|
degenerative spermatocyte homolog 2, lipid desaturase (Drosophila)
|
|
chr1_-_109651140
|
0.598
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr19_+_7607976
|
0.598
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr16_+_64958025
|
0.593
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr8_+_144522389
|
0.593
|
NM_052924
|
RHPN1
|
rhophilin, Rho GTPase binding protein 1
|
|
chr3_-_53855443
|
0.591
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr7_+_106292975
|
0.590
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr16_-_87300308
|
0.589
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr11_-_70971556
|
0.587
|
NM_001005405
|
KRTAP5-11
|
keratin associated protein 5-11
|
|
chr2_+_171280944
|
0.582
|
|
SP5
|
Sp5 transcription factor
|
|
chr20_+_42463419
|
0.581
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr8_+_8597075
|
0.579
|
NM_194284
|
CLDN23
|
claudin 23
|
|
chr17_-_44058612
|
0.574
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr9_-_14683280
|
0.573
|
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr8_-_140784416
|
0.573
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr17_-_7106987
|
0.571
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr3_-_168580764
|
0.571
|
NM_024687
|
ZBBX
|
zinc finger, B-box domain containing
|
|
chr4_+_2390469
|
0.571
|
NM_001193282
|
LOC402160
|
hypothetical protein LOC402160
|
|
chr13_-_69580459
|
0.570
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr14_+_71468902
|
0.570
|
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr7_-_126680372
|
0.568
|
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr7_+_106292932
|
0.567
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr7_-_11838260
|
0.567
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr3_+_16901455
|
0.567
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr22_-_35835548
|
0.565
|
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr2_+_170298700
|
0.565
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chrX_-_132947141
|
0.565
|
|
GPC3
|
glypican 3
|
|
chr3_-_47595190
|
0.562
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr12_-_7793335
|
0.561
|
NM_130441
NM_203503
|
CLEC4C
|
C-type lectin domain family 4, member C
|
|
chr1_-_27113018
|
0.561
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr22_-_20235372
|
0.560
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr5_+_99898922
|
0.557
|
NM_198507
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr1_+_246167115
|
0.557
|
NM_175911
|
OR2L13
|
olfactory receptor, family 2, subfamily L, member 13
|
|
chr7_-_150306169
|
0.554
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr14_-_26136799
|
0.550
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr17_+_5914364
|
0.547
|
NM_015253
|
WSCD1
|
WSC domain containing 1
|
|
chr1_-_40555413
|
0.547
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr18_-_6404900
|
0.546
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chrX_+_152562013
|
0.543
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr16_-_1862162
|
0.542
|
NM_001163560
|
C16orf73
|
chromosome 16 open reading frame 73
|
|
chr19_+_6723801
|
0.542
|
|
|
|
|
chr11_-_1576057
|
0.541
|
NM_001004325
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr17_+_42641426
|
0.540
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|