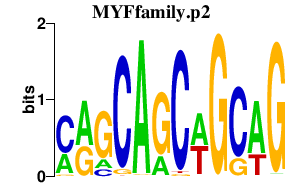
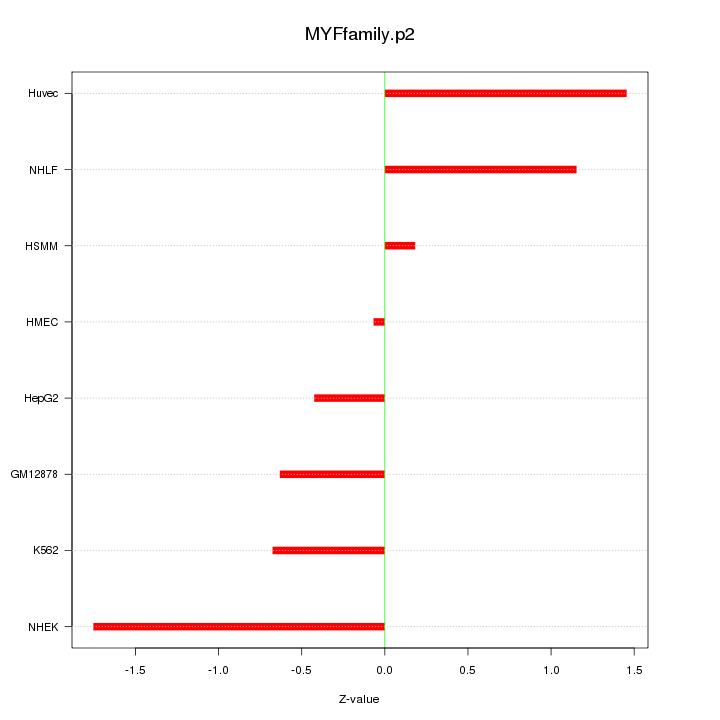
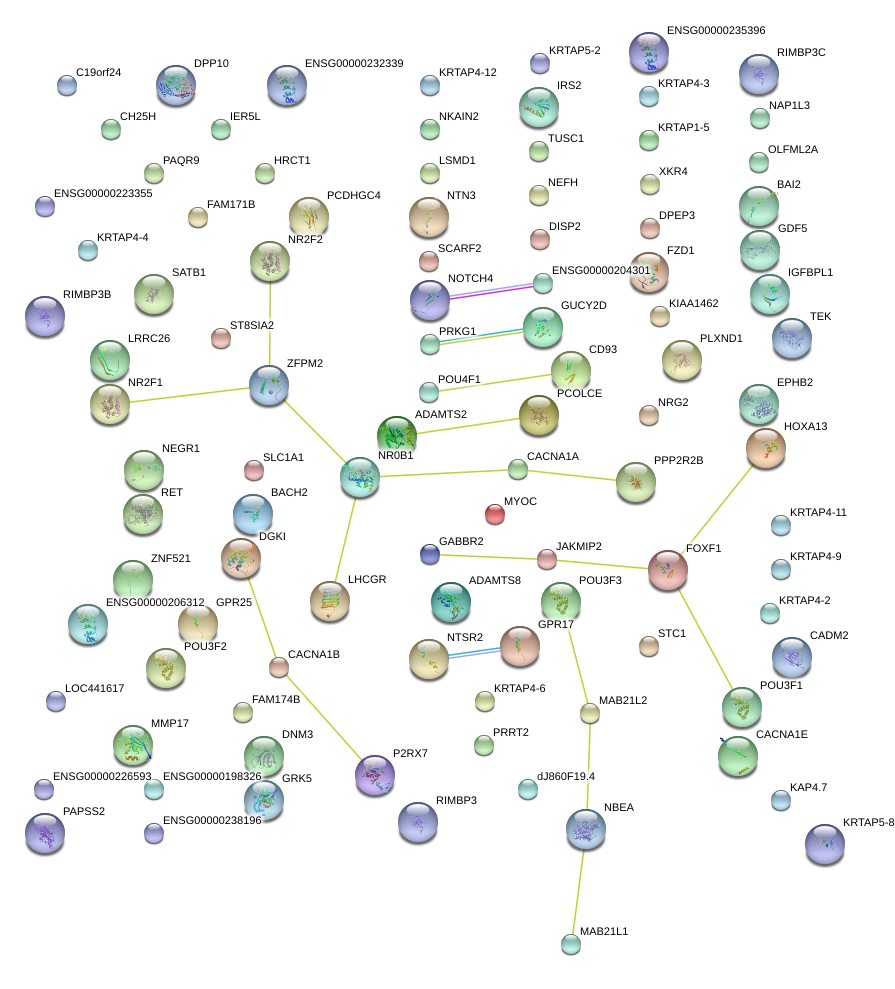
|
chr13_-_34948758
|
4.125
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr5_-_139402989
|
3.247
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr11_-_129803515
|
3.231
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr8_-_23768242
|
2.869
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr11_-_129803712
|
2.840
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr9_-_130980302
|
2.517
|
|
IER5L
|
immediate early response 5-like
|
|
chr2_-_48836313
|
2.382
|
NM_000233
|
LHCGR
|
luteinizing hormone/choriogonadotropin receptor
|
|
chr13_-_109236897
|
2.306
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr17_+_36493927
|
2.260
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr18_-_21186107
|
2.253
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr9_-_130980359
|
2.192
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr20_-_23014828
|
2.137
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr18_-_21185968
|
2.132
|
|
ZNF521
|
zinc finger protein 521
|
|
chr17_-_36533903
|
1.961
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr17_-_36577949
|
1.893
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr5_-_178704934
|
1.883
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr6_+_99389318
|
1.831
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr22_+_20068039
|
1.831
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr6_-_91063181
|
1.749
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr10_-_90957028
|
1.737
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr5_-_146238336
|
1.710
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_-_72520735
|
1.693
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr3_-_130807865
|
1.650
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr6_+_124166767
|
1.641
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr13_-_78075683
|
1.627
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr16_+_29731058
|
1.607
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chrX_-_92815222
|
1.605
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr17_-_36528113
|
1.591
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr15_-_90999606
|
1.591
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr9_-_38414422
|
1.587
|
NM_001007563
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1
|
|
chr1_-_169888373
|
1.575
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr10_+_89409332
|
1.563
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr8_+_56177567
|
1.560
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr3_+_85090721
|
1.536
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr6_-_32299757
|
1.532
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr20_-_33489383
|
1.510
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr3_-_18441763
|
1.492
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr16_+_85101605
|
1.479
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr20_+_2743613
|
1.470
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr16_+_85101734
|
1.453
|
|
FOXF1
|
forkhead box F1
|
|
chr15_+_94674849
|
1.439
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr8_+_106400322
|
1.374
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr1_-_32002222
|
1.366
|
NM_001703
|
BAI2
|
brain-specific angiogenesis inhibitor 2
|
|
chr2_+_187266899
|
1.328
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr12_+_130878870
|
1.309
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr10_-_30356344
|
1.303
|
|
KIAA1462
|
KIAA1462
|
|
chr19_-_13478037
|
1.281
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr5_+_140844810
|
1.263
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr9_+_4480193
|
1.257
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr7_-_27206249
|
1.226
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr16_+_2461500
|
1.221
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr11_+_74629597
|
1.205
|
NM_001195528
|
LOC441617
|
hypothetical LOC441617
|
|
chr10_+_120957084
|
1.197
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr12_+_120055013
|
1.197
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr7_-_137182125
|
1.190
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr15_+_90738108
|
1.188
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr15_+_38437703
|
1.185
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr17_-_7701465
|
1.184
|
|
LSMD1
|
LSM domain containing 1
|
|
chrX_-_30237403
|
1.179
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr9_+_35896188
|
1.165
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr5_-_147142457
|
1.163
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr17_-_36436979
|
1.159
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr1_+_170077234
|
1.154
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr9_-_25668230
|
1.152
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr16_-_66571863
|
1.144
|
NM_001129758
NM_022357
|
DPEP3
|
dipeptidase 3
|
|
chr13_+_34948885
|
1.144
|
|
NBEA
|
neurobeachin
|
|
chr1_+_22909916
|
1.141
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr1_-_72521079
|
1.135
|
|
|
|
|
chr22_-_19122047
|
1.133
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr22_+_28206180
|
1.131
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr9_-_139184247
|
1.131
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr2_+_114916368
|
1.116
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr9_-_66233945
|
1.108
|
|
|
|
|
chr10_+_42892506
|
1.107
|
NM_020630
NM_020975
|
RET
|
ret proto-oncogene
|
|
chr17_+_7846636
|
1.105
|
NM_000180
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific)
|
|
chr9_-_100510657
|
1.092
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr9_+_126579203
|
1.083
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr9_+_27099138
|
1.065
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr7_+_100037817
|
1.060
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr3_-_62835601
|
1.049
|
|
|
|
|
chr1_+_199108705
|
1.048
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr3_-_144164867
|
1.048
|
NM_198504
|
PAQR9
|
progestin and adipoQ receptor family member IX
|
|
chr2_-_11727705
|
1.045
|
NM_012344
|
NTSR2
|
neurotensin receptor 2
|
|
chr10_+_52504239
|
1.038
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr7_+_90731671
|
1.030
|
NM_003505
|
FZD1
|
frizzled homolog 1 (Drosophila)
|
|
chr7_+_137182230
|
1.029
|
|
|
|
|
chr11_-_1576057
|
1.026
|
NM_001004325
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr2_+_128119908
|
1.023
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr19_+_1226510
|
1.009
|
NM_017914
|
C19orf24
|
chromosome 19 open reading frame 24
|
|
chr12_-_53099277
|
1.002
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr11_+_70915960
|
1.001
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr11_+_65414677
|
0.996
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr19_+_47509274
|
0.973
|
NM_173633
|
TMEM145
|
transmembrane protein 145
|
|
chr14_+_58174489
|
0.970
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr5_-_147142368
|
0.968
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr12_-_53099269
|
0.968
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr5_-_147142444
|
0.966
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_-_87122936
|
0.956
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr12_+_4788602
|
0.950
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr9_+_76302071
|
0.945
|
NM_006914
|
RORB
|
RAR-related orphan receptor B
|
|
chr4_-_13155135
|
0.943
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr8_-_68821131
|
0.942
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr12_+_6915698
|
0.941
|
|
|
|
|
chr5_-_147142249
|
0.940
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr19_+_56319948
|
0.936
|
NM_001198558
NM_014441
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9
|
|
chrX_+_12066505
|
0.932
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr14_+_28306633
|
0.925
|
|
FOXG1
|
forkhead box G1
|
|
chr5_-_178705036
|
0.924
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr10_+_115793795
|
0.922
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr1_-_2451543
|
0.916
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr12_-_53099203
|
0.911
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_83830710
|
0.902
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr22_+_29333069
|
0.901
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr6_+_45498262
|
0.900
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_-_53367246
|
0.899
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr13_-_71338558
|
0.898
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr5_+_128823904
|
0.890
|
NM_133638
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19
|
|
chr15_+_39008822
|
0.890
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr5_+_126654354
|
0.863
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr7_-_27190882
|
0.862
|
|
HOXA11
|
homeobox A11
|
|
chr8_+_65656164
|
0.860
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr10_-_79067476
|
0.850
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr17_+_58908152
|
0.849
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr1_-_102235377
|
0.845
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr12_+_50342814
|
0.839
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr9_+_86474393
|
0.839
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr2_-_74498351
|
0.838
|
NM_001145054
|
C2orf81
|
chromosome 2 open reading frame 81
|
|
chr2_-_216944911
|
0.836
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr14_+_23590939
|
0.831
|
NM_138360
|
LRRC16B
|
leucine rich repeat containing 16B
|
|
chr20_-_47618059
|
0.831
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr17_+_1906262
|
0.825
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr11_-_106394039
|
0.820
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr3_-_191321601
|
0.815
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr5_+_137829065
|
0.812
|
|
EGR1
|
early growth response 1
|
|
chr2_+_100087812
|
0.806
|
|
|
|
|
chr7_+_119700924
|
0.804
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr1_+_22842704
|
0.804
|
NM_001114101
NM_172369
|
C1QC
|
complement component 1, q subcomponent, C chain
|
|
chr5_-_99898788
|
0.803
|
|
|
|
|
chr10_+_71833905
|
0.798
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr2_-_217267742
|
0.798
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr3_-_170864111
|
0.797
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr2_-_39040981
|
0.792
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr15_+_45796802
|
0.788
|
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_-_139992621
|
0.779
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr10_-_21845721
|
0.778
|
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr2_+_185171480
|
0.775
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr3_+_112273352
|
0.771
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr2_-_172675480
|
0.771
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr1_+_109810743
|
0.770
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr6_+_96570565
|
0.769
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr4_+_134292630
|
0.767
|
|
PCDH10
|
protocadherin 10
|
|
chr4_-_142273881
|
0.765
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr19_-_15204194
|
0.763
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr2_+_79593567
|
0.761
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr8_+_104452877
|
0.761
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr16_+_29730909
|
0.760
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr9_+_33667169
|
0.759
|
|
|
|
|
chr1_+_219119623
|
0.759
|
|
HLX
|
H2.0-like homeobox
|
|
chr8_+_99508425
|
0.757
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr17_-_74433048
|
0.752
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr5_+_129268352
|
0.749
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr1_-_38285033
|
0.747
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr9_+_101623957
|
0.742
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chrX_+_21302317
|
0.740
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr10_+_63803921
|
0.738
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr15_+_89229275
|
0.736
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr16_-_87450849
|
0.735
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr3_-_73756668
|
0.735
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr15_+_89228668
|
0.733
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr1_-_67163001
|
0.730
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr14_-_59407103
|
0.724
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr19_-_52426055
|
0.720
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr16_+_66433882
|
0.719
|
|
THAP11
|
THAP domain containing 11
|
|
chr9_-_131422842
|
0.716
|
NM_199350
|
C9orf50
|
chromosome 9 open reading frame 50
|
|
chr20_-_61462960
|
0.708
|
NM_000744
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4
|
|
chr13_+_109757593
|
0.704
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr15_+_45797977
|
0.704
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr2_-_219633481
|
0.702
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr3_+_3816079
|
0.701
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr7_+_39956396
|
0.699
|
|
|
|
|
chr17_-_36456949
|
0.698
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr3_-_124650081
|
0.697
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr11_-_132907499
|
0.696
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr8_+_37672427
|
0.695
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr7_+_103756339
|
0.694
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr12_+_101876151
|
0.692
|
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr14_-_50631407
|
0.684
|
|
TRIM9
|
tripartite motif containing 9
|
|
chrX_+_153767803
|
0.679
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr9_+_136673534
|
0.673
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr14_-_50631794
|
0.671
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr7_+_27190688
|
0.669
|
|
|
|
|
chr13_+_34414390
|
0.669
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr21_+_46226072
|
0.668
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr14_-_105851580
|
0.666
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr8_-_134156437
|
0.665
|
|
SLA
|
Src-like-adaptor
|
|
chr6_+_26291936
|
0.665
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr17_+_11085464
|
0.659
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr4_-_24641404
|
0.658
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr8_-_140784416
|
0.656
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr15_-_41698251
|
0.656
|
NM_153700
|
STRC
|
stereocilin
|