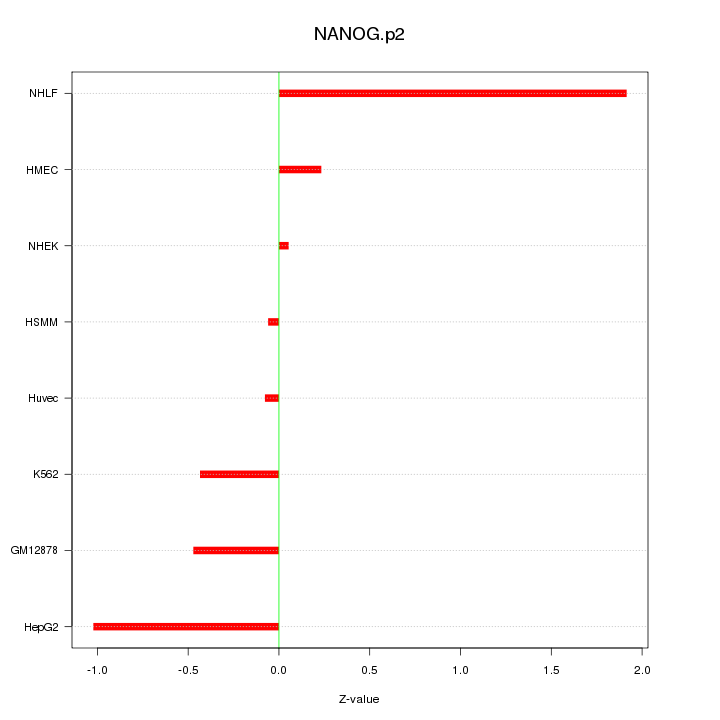
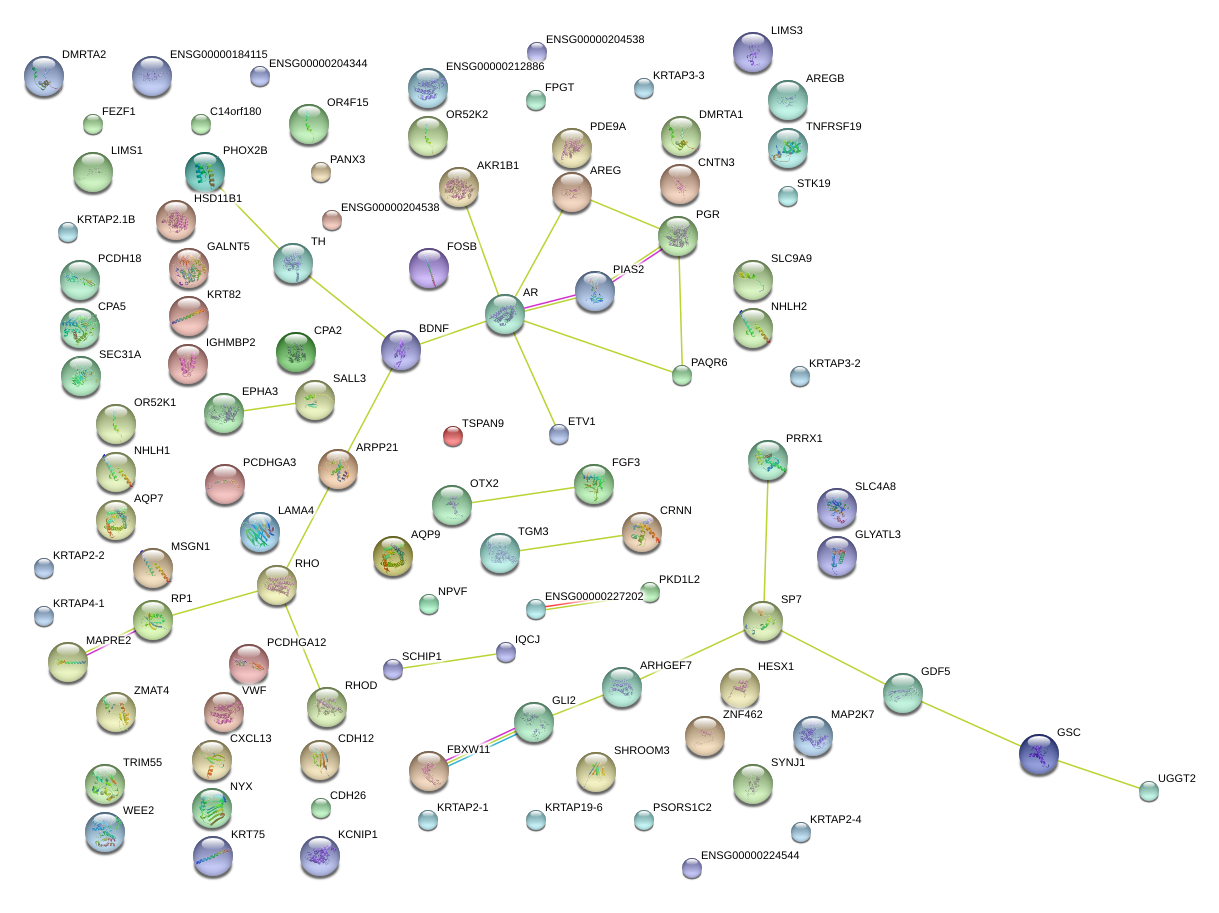
|
chr4_+_77575276
|
1.790
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr18_+_74841262
|
1.120
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr3_-_74652952
|
1.025
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr1_-_150653351
|
0.991
|
NM_016190
|
CRNN
|
cornulin
|
|
chr20_-_33489383
|
0.939
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr4_-_138673014
|
0.929
|
|
PCDH18
|
protocadherin 18
|
|
chr3_-_57209319
|
0.902
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr12_-_51114372
|
0.900
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr6_-_31215089
|
0.896
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr3_+_161040343
|
0.889
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_123986661
|
0.867
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr2_+_17861266
|
0.858
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr17_-_36403910
|
0.757
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr14_-_94306104
|
0.739
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr1_+_168898898
|
0.716
|
|
PRRX1
|
paired related homeobox 1
|
|
chr11_-_2149591
|
0.692
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr7_+_129693938
|
0.688
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr1_-_154484341
|
0.683
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr15_+_56217686
|
0.663
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr11_-_27637771
|
0.636
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_-_116185215
|
0.633
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr7_-_13992351
|
0.627
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr17_-_36456949
|
0.624
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr21_-_30836051
|
0.622
|
NM_181612
|
KRTAP19-6
|
keratin associated protein 19-6
|
|
chr12_-_52016270
|
0.606
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr14_-_56342097
|
0.603
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr11_+_4427100
|
0.584
|
NM_001005172
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2
|
|
chr8_-_40874485
|
0.575
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chrX_+_66705407
|
0.574
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr7_+_141054621
|
0.572
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr14_+_104117100
|
0.562
|
NM_001008404
|
C14orf180
|
chromosome 14 open reading frame 180
|
|
chr20_+_58004811
|
0.559
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chr20_+_2224612
|
0.554
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr16_-_79811475
|
0.545
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr5_+_140790141
|
0.538
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr3_+_89239494
|
0.536
|
|
EPHA3
|
EPH receptor A3
|
|
chr22_+_44828338
|
0.535
|
|
|
|
|
chr7_-_13996166
|
0.533
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr2_+_121271336
|
0.522
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr12_-_6103945
|
0.521
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr12_-_51086330
|
0.506
|
NM_033033
|
KRT82
|
keratin 82
|
|
chr7_+_129771865
|
0.504
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr7_-_121731721
|
0.503
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr1_-_50661716
|
0.501
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr4_-_138673078
|
0.500
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr5_-_22248630
|
0.497
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr2_+_157822355
|
0.496
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr17_-_19290745
|
0.496
|
|
|
|
|
chr5_-_168204430
|
0.495
|
|
|
|
|
chr3_+_161040439
|
0.479
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_-_145049853
|
0.478
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr8_+_55691179
|
0.474
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr1_+_74473672
|
0.472
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr3_+_130730171
|
0.468
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr4_+_78651929
|
0.461
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr3_+_35697432
|
0.459
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr8_+_67201684
|
0.450
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr4_-_41445693
|
0.444
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr6_+_44076281
|
0.442
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr19_+_7874701
|
0.441
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr2_-_110665188
|
0.432
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr21_+_43024936
|
0.430
|
|
PDE9A
|
phosphodiesterase 9A
|
|
chr13_+_23042722
|
0.428
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chrX_+_41191630
|
0.426
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr6_+_49575629
|
0.423
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr9_+_108665168
|
0.423
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr5_+_140703784
|
0.421
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr15_+_100175888
|
0.413
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr1_+_207944807
|
0.411
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr17_-_36594652
|
0.408
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr5_+_169713458
|
0.408
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr7_-_25234582
|
0.407
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr19_+_50663089
|
0.407
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr11_-_69343128
|
0.406
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr6_-_112682401
|
0.406
|
|
LAMA4
|
laminin, alpha 4
|
|
chr1_+_109884016
|
0.394
|
NM_031936
|
GPR61
|
G protein-coupled receptor 61
|
|
chr5_-_35974493
|
0.394
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr11_-_57173823
|
0.392
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr9_+_135491305
|
0.389
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr2_+_27155197
|
0.386
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr3_-_113842652
|
0.382
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_+_140790310
|
0.381
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr8_+_32698892
|
0.380
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr13_-_31784088
|
0.380
|
NM_001136571
|
ZAR1L
|
zygote arrest 1-like
|
|
chr9_-_132759045
|
0.371
|
NM_198180
|
QRFP
|
pyroglutamylated RFamide peptide
|
|
chr6_-_169392771
|
0.369
|
|
|
|
|
chr5_+_140790365
|
0.368
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_+_180836050
|
0.367
|
|
|
|
|
chr6_-_112682573
|
0.366
|
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_112682459
|
0.365
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr13_+_100902942
|
0.357
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr7_+_150376537
|
0.357
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr17_-_36418869
|
0.356
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr10_-_105835525
|
0.356
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr5_+_140848905
|
0.355
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr10_-_13789628
|
0.354
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chrX_-_21586337
|
0.353
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr6_+_34314545
|
0.352
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr4_-_26100986
|
0.350
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr3_-_116272911
|
0.346
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_102522093
|
0.345
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr22_-_39189369
|
0.343
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr3_+_16191159
|
0.343
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr19_+_3713650
|
0.337
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr2_+_225973845
|
0.335
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr21_-_30460841
|
0.333
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr14_-_104330982
|
0.333
|
NM_005163
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr20_+_43691797
|
0.332
|
NM_080753
|
WFDC10A
|
WAP four-disulfide core domain 10A
|
|
chr12_-_90100710
|
0.327
|
|
DCN
|
decorin
|
|
chr5_+_170668892
|
0.327
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr14_+_51025604
|
0.322
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr6_-_39801088
|
0.320
|
NM_145027
|
KIF6
|
kinesin family member 6
|
|
chr17_-_9635325
|
0.320
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr10_-_93659164
|
0.319
|
NM_152429
|
FGFBP3
|
fibroblast growth factor binding protein 3
|
|
chr6_+_134800533
|
0.316
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr3_-_121445831
|
0.314
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chrX_+_37777730
|
0.313
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr4_+_110968594
|
0.313
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr2_-_27967458
|
0.312
|
|
LOC100302650
|
hypothetical LOC100302650
|
|
chr8_-_134156353
|
0.310
|
|
SLA
|
Src-like-adaptor
|
|
chr2_+_210152609
|
0.308
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_-_40510923
|
0.308
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr12_-_51332214
|
0.307
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr5_+_140790510
|
0.304
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr8_-_142333339
|
0.301
|
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr3_+_150675117
|
0.300
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr8_-_93176819
|
0.300
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_-_41445479
|
0.299
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr5_+_140494971
|
0.299
|
NM_015669
|
PCDHB5
|
protocadherin beta 5
|
|
chr5_+_140460240
|
0.298
|
NM_018937
|
PCDHB3
|
protocadherin beta 3
|
|
chr8_-_20084892
|
0.297
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr17_-_39094787
|
0.296
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr10_+_118073929
|
0.296
|
NM_198515
|
C10orf96
|
chromosome 10 open reading frame 96
|
|
chr2_+_161981138
|
0.294
|
|
TBR1
|
T-box, brain, 1
|
|
chr9_+_6205804
|
0.290
|
|
IL33
|
interleukin 33
|
|
chr7_+_113842470
|
0.289
|
|
FOXP2
|
forkhead box P2
|
|
chr2_-_224175339
|
0.289
|
|
SCG2
|
secretogranin II
|
|
chr11_-_83071084
|
0.287
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_+_207823667
|
0.287
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr12_-_89922888
|
0.285
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr7_-_41709191
|
0.283
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr12_+_111713931
|
0.282
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr8_-_93176618
|
0.282
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_140509803
|
0.280
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr1_-_155336223
|
0.278
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr7_+_143437539
|
0.276
|
NM_001005480
|
OR2A2
|
olfactory receptor, family 2, subfamily A, member 2
|
|
chr13_+_108079570
|
0.275
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr2_+_95326753
|
0.275
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr2_+_96269703
|
0.272
|
NM_001037228
|
LOC285033
|
hypothetical protein LOC285033
|
|
chr17_-_24972471
|
0.272
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr8_-_134156437
|
0.271
|
|
SLA
|
Src-like-adaptor
|
|
chr3_-_21767819
|
0.270
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr15_-_91078307
|
0.269
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr7_-_5429702
|
0.269
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr11_-_104421240
|
0.269
|
NM_001017534
NM_052889
|
CARD16
CASP1
|
caspase recruitment domain family, member 16
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr8_+_24354387
|
0.267
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr12_+_111714096
|
0.264
|
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr10_+_26545241
|
0.263
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr1_-_32036802
|
0.262
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr2_-_224175386
|
0.261
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr14_+_75907442
|
0.259
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr14_+_77939845
|
0.257
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr11_-_128217550
|
0.257
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr1_+_151456683
|
0.255
|
NM_001195571
|
PRR9
|
proline rich 9
|
|
chr1_-_158098962
|
0.255
|
NM_001013661
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
|
chr11_+_63775567
|
0.250
|
NM_000932
NM_001184883
|
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific)
|
|
chr5_+_176744037
|
0.246
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr2_+_1396239
|
0.245
|
NM_000547
NM_175719
|
TPO
|
thyroid peroxidase
|
|
chrX_+_50044279
|
0.245
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr5_+_31229609
|
0.245
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr11_-_57792206
|
0.241
|
NM_207374
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1
|
|
chr1_-_53380709
|
0.240
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr2_+_113542017
|
0.240
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chrX_-_108611940
|
0.238
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr6_-_32605898
|
0.237
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr5_-_24535386
|
0.237
|
NM_001190450
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr12_+_25096472
|
0.236
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr9_-_112381593
|
0.236
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chrX_+_102918409
|
0.235
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr6_+_84619703
|
0.234
|
NM_001009994
|
RIPPLY2
|
ripply2 homolog (zebrafish)
|
|
chr2_+_161980865
|
0.234
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr17_+_39990124
|
0.233
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr6_+_29902734
|
0.233
|
NM_002127
|
HLA-G
|
major histocompatibility complex, class I, G
|
|
chr20_+_42463279
|
0.232
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr16_-_66075216
|
0.231
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr19_+_59333287
|
0.231
|
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chrX_+_48978870
|
0.228
|
NM_014008
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr17_-_36528113
|
0.227
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr12_-_90100733
|
0.225
|
NM_001920
|
DCN
|
decorin
|
|
chr3_+_69895310
|
0.225
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr13_+_48178620
|
0.224
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr7_+_94135384
|
0.224
|
|
PEG10
|
paternally expressed 10
|
|
chr12_+_50342814
|
0.223
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr1_-_230717850
|
0.223
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr12_-_107551783
|
0.222
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr4_+_114190233
|
0.221
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr12_+_25096755
|
0.221
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr4_-_174687931
|
0.220
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chrX_+_135058402
|
0.220
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_-_99555399
|
0.219
|
NM_001190415
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa
|