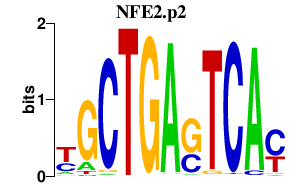
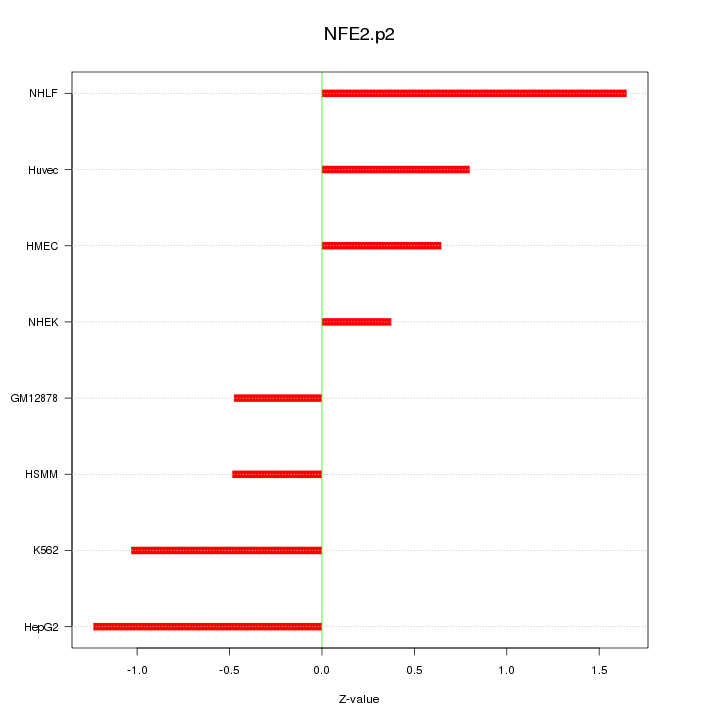
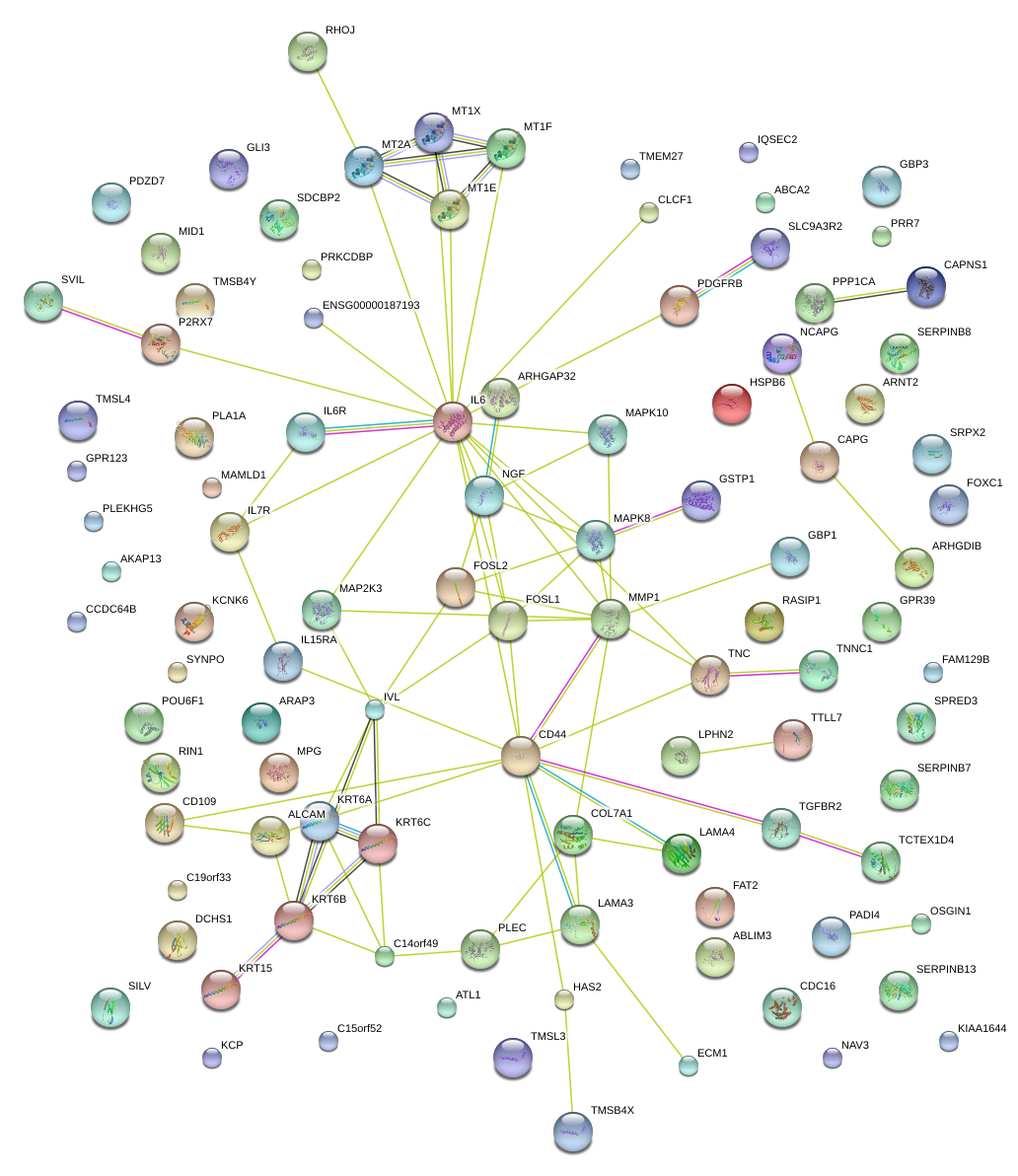
|
chr5_-_141041982
|
1.337
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr12_+_76749199
|
1.317
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr5_+_150000394
|
1.218
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr5_-_141041933
|
1.107
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr11_-_6298284
|
0.937
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr5_+_35892747
|
0.883
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr14_+_62740878
|
0.841
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chrX_+_99786044
|
0.809
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr2_-_85494639
|
0.778
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr11_-_102174102
|
0.775
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr9_-_116920232
|
0.775
|
NM_002160
|
TNC
|
tenascin C
|
|
chr18_+_59405513
|
0.754
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr10_-_102780840
|
0.752
|
|
PDZD7
|
PDZ domain containing 7
|
|
chr11_-_102174032
|
0.726
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr7_-_42243142
|
0.716
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr10_-_30064600
|
0.708
|
NM_003174
|
SVIL
|
supervillin
|
|
chrX_-_15593074
|
0.706
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr7_-_42243318
|
0.696
|
|
GLI3
|
GLI family zinc finger 3
|
|
chrX_+_12903145
|
0.693
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr11_-_65860450
|
0.693
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr11_-_66898223
|
0.674
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr10_-_88720908
|
0.665
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chrX_-_10761729
|
0.662
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr19_+_43502380
|
0.659
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_+_43572679
|
0.656
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr11_-_6633427
|
0.644
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr3_-_48607596
|
0.629
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr16_+_2023279
|
0.618
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr2_-_85494664
|
0.604
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr1_-_45045543
|
0.601
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chrX_+_99785818
|
0.599
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr18_+_19706979
|
0.598
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr18_+_59788242
|
0.596
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr1_+_17507276
|
0.593
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr14_-_95011790
|
0.587
|
NM_152592
|
C14orf49
|
chromosome 14 open reading frame 49
|
|
chr11_+_67107641
|
0.586
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr15_-_38417564
|
0.574
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr16_-_3025542
|
0.555
|
NM_001103175
|
CCDC64B
|
coiled-coil domain containing 64B
|
|
chr20_-_1257782
|
0.549
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr1_-_89261021
|
0.547
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr17_+_21131940
|
0.534
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chrX_+_149282097
|
0.530
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr9_-_129381086
|
0.528
|
NM_001035534
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr2_+_132890616
|
0.526
|
NM_001508
|
GPR39
|
G protein-coupled receptor 39
|
|
chr7_+_22733290
|
0.521
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr5_-_149515613
|
0.519
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_+_148501178
|
0.517
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr16_+_82544327
|
0.516
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr10_+_134751398
|
0.512
|
NM_001083909
|
GPR123
|
G protein-coupled receptor 123
|
|
chr9_-_139042526
|
0.511
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr5_-_149515502
|
0.501
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_54646061
|
0.500
|
NM_006928
|
PMEL
|
premelanosome protein
|
|
chr1_-_6474346
|
0.493
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_-_53935648
|
0.485
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chr18_+_59593591
|
0.475
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr15_+_84021271
|
0.472
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr2_+_28469275
|
0.472
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr5_+_176807931
|
0.471
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr11_+_67107852
|
0.463
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr1_-_115682374
|
0.463
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr12_-_51153788
|
0.463
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr11_-_128399218
|
0.460
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr8_-_145097031
|
0.455
|
NM_201380
|
PLEC
|
plectin
|
|
chr1_+_152644471
|
0.448
|
|
IL6R
|
interleukin 6 receptor
|
|
chr19_-_40939756
|
0.442
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr3_+_106568334
|
0.442
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr1_+_81544432
|
0.442
|
|
LPHN2
|
latrophilin 2
|
|
chr1_+_151147646
|
0.441
|
NM_005547
|
IVL
|
involucrin
|
|
chr3_+_30622905
|
0.441
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr5_-_149515331
|
0.441
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_15005761
|
0.436
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr3_+_120799411
|
0.435
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr17_-_36928604
|
0.434
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr1_+_148747208
|
0.432
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr19_+_43486640
|
0.431
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr7_-_128330615
|
0.430
|
|
KCP
|
kielin/chordin-like protein
|
|
chrX_-_53327473
|
0.428
|
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr1_-_84237316
|
0.428
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr8_-_145088679
|
0.426
|
NM_201383
|
PLEC
|
plectin
|
|
chr22_-_43040063
|
0.425
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr12_-_49897743
|
0.424
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr4_-_87500398
|
0.423
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr19_+_41322841
|
0.422
|
|
CAPNS1
|
calpain, small subunit 1
|
|
chr6_+_1556543
|
0.421
|
|
FOXC1
|
forkhead box C1
|
|
chr16_+_55199978
|
0.416
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr6_+_74462630
|
0.416
|
|
CD109
|
CD109 molecule
|
|
chr11_+_35117226
|
0.416
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr15_+_78520634
|
0.413
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr11_-_65424434
|
0.412
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr10_-_6059460
|
0.410
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr8_-_122722674
|
0.405
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr4_+_40953698
|
0.404
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr11_+_47386621
|
0.403
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr9_+_35528890
|
0.401
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_-_18848713
|
0.400
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr2_+_28469172
|
0.399
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr12_-_54522881
|
0.397
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr3_+_11153778
|
0.395
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr12_-_90097384
|
0.394
|
NM_133503
|
DCN
|
decorin
|
|
chr16_+_56219802
|
0.391
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr9_-_139042416
|
0.391
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr3_+_49002285
|
0.389
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr19_+_41119861
|
0.389
|
NM_024509
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3
|
|
chr11_+_128068982
|
0.388
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr11_-_65424436
|
0.388
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_151788341
|
0.387
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr8_-_26780665
|
0.387
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr11_+_48303047
|
0.387
|
NM_001004702
|
OR4C3
|
olfactory receptor, family 4, subfamily C, member 3
|
|
chr16_+_88515917
|
0.386
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr7_+_100557098
|
0.386
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr1_+_148747110
|
0.385
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr3_-_58175437
|
0.383
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr11_-_65424385
|
0.383
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr21_-_35181956
|
0.382
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_106653429
|
0.381
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr17_+_58058479
|
0.379
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr15_+_72315719
|
0.379
|
NM_025055
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr7_+_1543644
|
0.379
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr15_+_99276982
|
0.378
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr22_-_43986896
|
0.377
|
|
KIAA0930
|
KIAA0930
|
|
chr21_-_35343331
|
0.376
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_-_73830480
|
0.375
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr6_+_74462654
|
0.370
|
|
CD109
|
CD109 molecule
|
|
chr15_-_68971665
|
0.369
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr6_+_41712907
|
0.366
|
|
MDFI
|
MyoD family inhibitor
|
|
chr17_-_7064092
|
0.365
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr12_-_46439127
|
0.364
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr17_-_7063625
|
0.363
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr7_+_90177082
|
0.362
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_+_74462226
|
0.361
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr1_-_161105174
|
0.359
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chr11_+_62136788
|
0.358
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr3_-_48576204
|
0.357
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr9_+_35528628
|
0.356
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_1536847
|
0.353
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr11_+_101488380
|
0.352
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr11_+_19328846
|
0.352
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr16_+_4370879
|
0.352
|
|
VASN
|
vasorin
|
|
chr2_-_219566364
|
0.352
|
NM_005209
NM_057093
NM_057094
|
CRYBA2
|
crystallin, beta A2
|
|
chr1_-_17180659
|
0.351
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr16_+_15503623
|
0.351
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr11_+_128069203
|
0.351
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr2_+_28469199
|
0.350
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr16_+_55684031
|
0.346
|
|
CPNE2
|
copine II
|
|
chr20_-_1257809
|
0.346
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr9_+_130942029
|
0.345
|
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr22_-_28972683
|
0.345
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr11_-_89248832
|
0.338
|
NM_001164397
|
TRIM64B
|
tripartite motif containing 64B
|
|
chr16_+_31274030
|
0.337
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chrX_-_19727828
|
0.337
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr9_-_101621964
|
0.337
|
|
|
|
|
chr19_-_3920736
|
0.336
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr14_-_104507862
|
0.335
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr3_-_115826481
|
0.335
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_77261401
|
0.333
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr19_+_12996368
|
0.330
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_-_176205672
|
0.329
|
NM_033127
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr1_+_152644292
|
0.328
|
NM_000565
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr18_+_41560915
|
0.327
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr1_+_36394338
|
0.324
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr18_+_59705918
|
0.323
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr3_+_106568243
|
0.323
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr16_-_2848106
|
0.322
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr17_-_35975227
|
0.321
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr11_+_47386707
|
0.320
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr21_-_35343448
|
0.320
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_-_34652629
|
0.319
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr14_+_22422270
|
0.318
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr13_+_52500830
|
0.317
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr4_-_68678181
|
0.317
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr9_-_35101534
|
0.316
|
|
KIAA1539
|
KIAA1539
|
|
chr10_+_115302767
|
0.315
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr10_-_99521701
|
0.314
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr17_-_35911340
|
0.311
|
NM_032865
|
TNS4
|
tensin 4
|
|
chrX_-_110400406
|
0.311
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr19_+_12996830
|
0.310
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr6_+_74462577
|
0.309
|
|
CD109
|
CD109 molecule
|
|
chr14_+_78815406
|
0.309
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr17_-_36877002
|
0.308
|
NM_002278
|
KRT32
|
keratin 32
|
|
chr1_+_27541086
|
0.308
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr17_+_18566060
|
0.307
|
NM_001037330
|
TRIM16L
|
tripartite motif containing 16-like
|
|
chr2_-_73170290
|
0.306
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr10_-_29963906
|
0.306
|
NM_021738
|
SVIL
|
supervillin
|
|
chr3_-_115825742
|
0.304
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_+_168381811
|
0.303
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr12_+_54611212
|
0.301
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr1_+_152644668
|
0.298
|
|
IL6R
|
interleukin 6 receptor
|
|
chr1_+_109827975
|
0.297
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chrX_+_47326594
|
0.295
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr11_+_35154775
|
0.293
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr7_-_1465487
|
0.291
|
|
MICALL2
|
MICAL-like 2
|
|
chr17_+_58058774
|
0.291
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr16_+_66128843
|
0.290
|
NM_001193524
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr17_-_31231421
|
0.290
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr11_+_5916033
|
0.288
|
|
|
|
|
chr21_-_35343065
|
0.287
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr7_-_98305599
|
0.284
|
NM_001134450
NM_001134451
NM_152913
|
TMEM130
|
transmembrane protein 130
|
|
chr21_+_46367216
|
0.284
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_-_53099269
|
0.283
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_-_187744808
|
0.282
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|