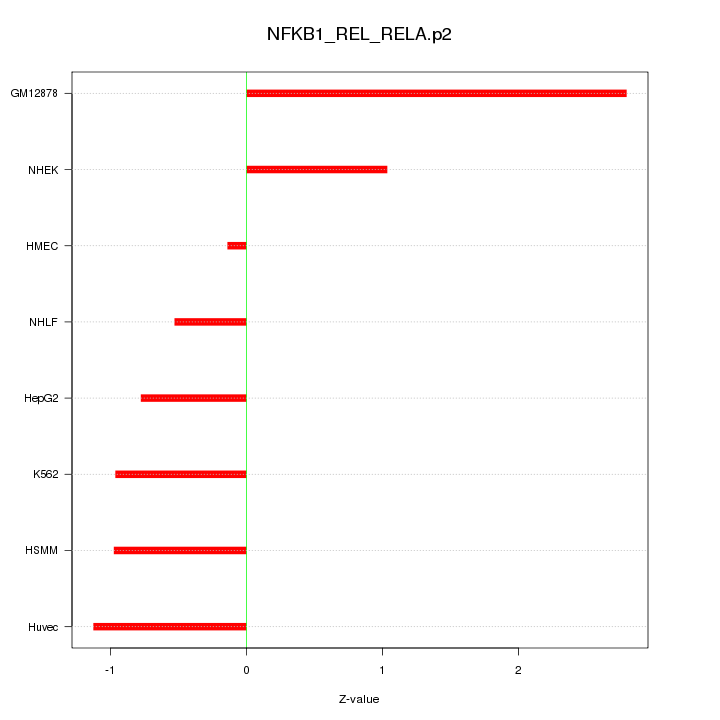
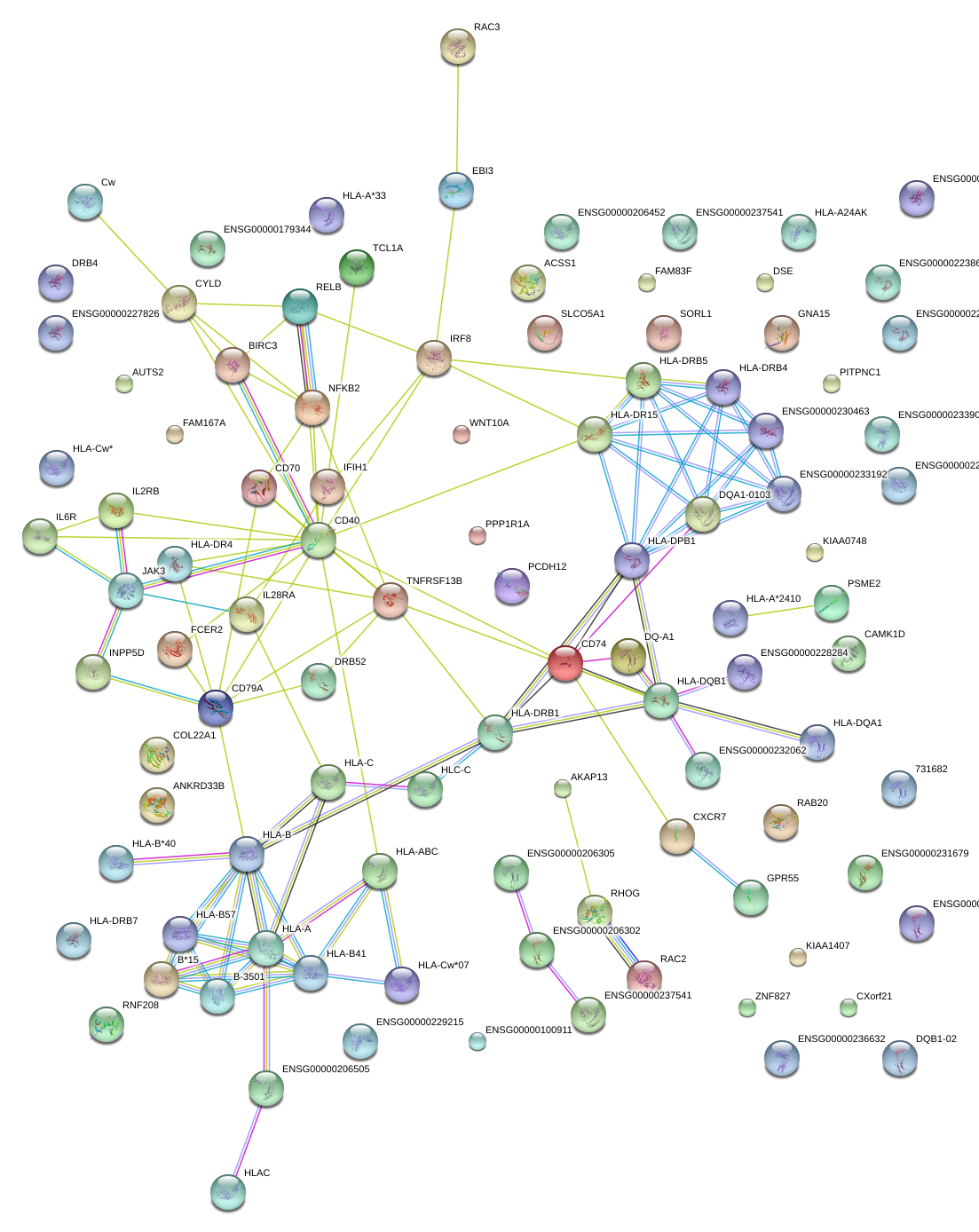
|
chr19_+_4180494
|
1.635
|
NM_005755
|
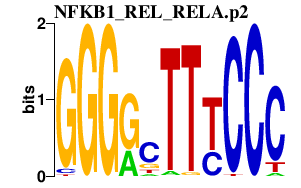
EBI3
|
Epstein-Barr virus induced 3
|
|
chr14_-_95250155
|
1.487
|
NM_001098725
NM_021966
|
TCL1A
|
T-cell leukemia/lymphoma 1A
|
|
chr19_+_50196526
|
1.303
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr10_+_12431714
|
1.224
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr10_+_104145421
|
1.128
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr8_-_70909754
|
1.080
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr17_-_16816113
|
1.041
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr2_+_219453487
|
1.022
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr10_+_104144218
|
1.016
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr22_-_35875890
|
0.958
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr16_+_84490267
|
0.945
|
NM_002163
|
IRF8
|
interferon regulatory factor 8
|
|
chr5_+_10617431
|
0.924
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr19_+_3087026
|
0.911
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr19_-_6542012
|
0.905
|
|
CD70
|
CD70 molecule
|
|
chr1_-_24386324
|
0.870
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr19_-_7672978
|
0.863
|
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr2_-_162883062
|
0.851
|
NM_022168
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr11_+_120828058
|
0.846
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr9_-_139235595
|
0.834
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr19_-_17819773
|
0.829
|
|
JAK3
|
Janus kinase 3
|
|
chr4_-_147079018
|
0.818
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr2_+_233633348
|
0.809
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr2_-_231498184
|
0.784
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr22_-_35970230
|
0.779
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr20_+_44180296
|
0.772
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr7_+_68702252
|
0.762
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr12_-_53268538
|
0.759
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr17_+_62804036
|
0.752
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr20_-_24986501
|
0.735
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr22_+_38720881
|
0.732
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr6_-_31432934
|
0.724
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr6_+_32713160
|
0.715
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr11_+_101693403
|
0.712
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr19_+_47073029
|
0.681
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr13_-_110011866
|
0.676
|
NM_017817
|
RAB20
|
RAB20, member RAS oncogene family
|
|
chr11_+_120828275
|
0.657
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr12_-_53661888
|
0.639
|
NM_001098815
|
KIAA0748
|
KIAA0748
|
|
chr8_-_11361594
|
0.633
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr2_+_237143118
|
0.630
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr5_-_141318810
|
0.629
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr20_+_44180355
|
0.621
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr2_+_233633279
|
0.618
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr1_+_152644668
|
0.617
|
|
IL6R
|
interleukin 6 receptor
|
|
chrX_-_30505804
|
0.614
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr15_+_83724743
|
0.609
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr19_-_6542082
|
0.600
|
|
CD70
|
CD70 molecule
|
|
chr14_-_23686269
|
0.591
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr3_-_115258053
|
0.591
|
NM_020817
|
KIAA1407
|
KIAA1407
|
|
chr16_+_49333461
|
0.591
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr16_+_49333513
|
0.591
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr10_+_12431477
|
0.589
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr6_+_116798791
|
0.587
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr8_-_139995417
|
0.571
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr6_-_32659958
|
0.564
|
|
HLA-DRB1
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 5
|
|
chr5_-_149772469
|
0.562
|
NM_001025159
NM_004355
NM_001025158
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr6_+_14225932
|
0.555
|
|
CD83
|
CD83 molecule
|
|
chr7_-_44985084
|
0.552
|
|
MYO1G
|
myosin IG
|
|
chr19_-_48976852
|
0.547
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr12_+_6925997
|
0.542
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr5_+_110587546
|
0.541
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr20_-_4743768
|
0.540
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr19_-_48977247
|
0.534
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr6_+_106640903
|
0.533
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_-_17819814
|
0.532
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr6_+_32919873
|
0.532
|
|
|
|
|
chr3_-_113334657
|
0.532
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr22_-_32646325
|
0.530
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr17_-_33178987
|
0.530
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr6_-_132764257
|
0.528
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr7_-_99899829
|
0.527
|
NM_001004323
|
C7orf61
|
chromosome 7 open reading frame 61
|
|
chrX_+_14457340
|
0.522
|
NM_001118885
NM_001118886
NM_001171942
NM_002063
|
GLRA2
|
glycine receptor, alpha 2
|
|
chr5_-_168660293
|
0.522
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr6_+_149109841
|
0.521
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr21_+_25933459
|
0.515
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr1_-_37873077
|
0.512
|
NM_001038633
|
RSPO1
|
R-spondin homolog (Xenopus laevis)
|
|
chrX_+_18353623
|
0.511
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chrX_+_130019896
|
0.511
|
NM_144967
|
ARHGAP36
|
Rho GTPase activating protein 36
|
|
chrX_+_12795111
|
0.506
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chrX_+_78087484
|
0.504
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr1_-_207892420
|
0.498
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr5_+_177473071
|
0.496
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr22_-_36212160
|
0.496
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr17_-_31231421
|
0.496
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr1_-_157003095
|
0.495
|
NM_001005185
|
OR6N1
|
olfactory receptor, family 6, subfamily N, member 1
|
|
chr19_-_4289750
|
0.487
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr1_-_1614005
|
0.485
|
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chr15_+_79376295
|
0.485
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr13_+_52500830
|
0.482
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr4_-_174327377
|
0.482
|
|
|
|
|
chr12_+_6926014
|
0.477
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr19_+_43502301
|
0.477
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr1_-_85235210
|
0.476
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr22_-_35970133
|
0.476
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr2_-_235070431
|
0.474
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chrX_-_111970662
|
0.472
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr1_-_153107084
|
0.471
|
|
|
|
|
chr13_+_23051426
|
0.470
|
NM_018647
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr11_+_66580884
|
0.470
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr11_+_66580861
|
0.468
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr4_+_103641762
|
0.467
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr1_+_40193424
|
0.466
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr14_+_102313566
|
0.465
|
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr8_+_73612179
|
0.459
|
NM_004770
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2
|
|
chr17_+_26742903
|
0.458
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr1_-_164402700
|
0.458
|
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr11_+_65740789
|
0.457
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr19_+_17723285
|
0.457
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr3_-_58175437
|
0.452
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr1_-_207891264
|
0.451
|
NM_001017402
|
LAMB3
|
laminin, beta 3
|
|
chr1_-_200396354
|
0.450
|
NM_002832
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr17_+_60563917
|
0.450
|
NM_001081955
NM_001165933
NM_003835
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr2_+_95055155
|
0.447
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr15_+_71131877
|
0.446
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr10_-_134940315
|
0.446
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr5_-_149772563
|
0.445
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr19_-_4133495
|
0.439
|
NM_001193285
NM_016539
|
SIRT6
|
sirtuin 6
|
|
chr15_+_74139325
|
0.436
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chrX_+_103243762
|
0.431
|
NM_001143978
|
ZCCHC18
|
zinc finger, CCHC domain containing 18
|
|
chr1_-_164402575
|
0.428
|
NM_001017961
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr7_+_68702516
|
0.428
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr11_-_57092332
|
0.425
|
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr9_-_131555111
|
0.425
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr6_+_106640881
|
0.424
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_+_68815416
|
0.423
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr19_+_43502591
|
0.423
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr17_-_59172852
|
0.423
|
|
STRADA
|
STE20-related kinase adaptor alpha
|
|
chr12_-_56447090
|
0.422
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr4_+_103641854
|
0.422
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr14_-_23106779
|
0.422
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr1_-_180840016
|
0.421
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr17_+_69974116
|
0.420
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chr10_-_105427898
|
0.419
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr8_-_11096257
|
0.419
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr19_-_44427446
|
0.418
|
NM_172139
|
IL28B
|
interleukin 28B (interferon, lambda 3)
|
|
chr17_-_36451189
|
0.418
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr17_+_60563963
|
0.411
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr6_+_31648071
|
0.409
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr17_+_43328514
|
0.409
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr7_-_44985202
|
0.409
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr4_+_103641511
|
0.408
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr14_+_102313585
|
0.404
|
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr11_-_57092028
|
0.398
|
NM_198183
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr1_-_27834210
|
0.398
|
NM_005248
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr2_+_228386800
|
0.397
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr13_+_44813479
|
0.397
|
|
LOC100190939
|
hypothetical LOC100190939
|
|
chr19_+_2427122
|
0.396
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr1_+_152644471
|
0.394
|
|
IL6R
|
interleukin 6 receptor
|
|
chr19_-_11549441
|
0.394
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_-_64040072
|
0.388
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr19_+_1018536
|
0.387
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr16_+_49288550
|
0.386
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr18_+_22060383
|
0.384
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr6_-_32919751
|
0.382
|
NM_148919
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr12_+_129212955
|
0.381
|
NM_007197
|
FZD10
|
frizzled homolog 10 (Drosophila)
|
|
chr16_+_55580910
|
0.379
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr2_-_74522563
|
0.377
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr15_-_72804763
|
0.375
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr5_-_171547844
|
0.374
|
|
STK10
|
serine/threonine kinase 10
|
|
chr9_-_138236775
|
0.373
|
NM_178138
|
LHX3
|
LIM homeobox 3
|
|
chr1_+_152644292
|
0.371
|
NM_000565
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr12_-_51476047
|
0.371
|
NM_057088
|
KRT3
|
keratin 3
|
|
chr3_+_123279439
|
0.369
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr20_+_29526751
|
0.369
|
NM_014012
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1
|
|
chr7_+_150376537
|
0.364
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr19_-_46551670
|
0.364
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr17_+_35728039
|
0.361
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr17_-_7531456
|
0.361
|
NM_000546
NM_001126112
NM_001126113
NM_001126114
|
TP53
|
tumor protein p53
|
|
chr17_-_23151601
|
0.360
|
NM_000625
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr11_+_128266460
|
0.360
|
NM_000890
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5
|
|
chr5_-_171547700
|
0.357
|
|
STK10
|
serine/threonine kinase 10
|
|
chr19_+_54786723
|
0.356
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr10_-_133645301
|
0.353
|
NM_004052
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chrX_+_28515479
|
0.353
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr17_+_63713171
|
0.353
|
|
|
|
|
chr6_-_33016561
|
0.352
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr5_+_169465478
|
0.351
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr19_-_55071645
|
0.351
|
|
AKT1S1
|
AKT1 substrate 1 (proline-rich)
|
|
chr1_-_37872874
|
0.350
|
|
RSPO1
|
R-spondin homolog (Xenopus laevis)
|
|
chr1_-_158182006
|
0.350
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr1_+_159959072
|
0.345
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr14_+_69148066
|
0.345
|
|
KIAA0247
|
KIAA0247
|
|
chr1_-_202730135
|
0.343
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr11_-_76800525
|
0.339
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr14_+_18623364
|
0.339
|
NM_001005356
|
POTEG
|
POTE ankyrin domain family, member G
|
|
chr19_-_6542136
|
0.338
|
NM_001252
|
CD70
|
CD70 molecule
|
|
chr7_-_44985175
|
0.337
|
|
MYO1G
|
myosin IG
|
|
chr20_-_62181231
|
0.337
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr5_+_167651063
|
0.336
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr6_-_31246159
|
0.335
|
NM_002701
|
POU5F1
POU5F1B
POU5F1P3
|
POU class 5 homeobox 1
POU class 5 homeobox 1B
POU class 5 homeobox 1 pseudogene 3
|
|
chr17_-_1874927
|
0.335
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr15_+_57517564
|
0.335
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr17_+_35727998
|
0.335
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr17_+_4795128
|
0.334
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr14_-_23878871
|
0.332
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr12_-_108231407
|
0.332
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr5_-_171547925
|
0.330
|
NM_005990
|
STK10
|
serine/threonine kinase 10
|
|
chr6_-_143307976
|
0.327
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr16_-_73798539
|
0.322
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr7_+_143402207
|
0.322
|
NM_001004488
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25
|
|
chr9_+_35739321
|
0.320
|
|
RGP1
|
RGP1 retrograde golgi transport homolog (S. cerevisiae)
|