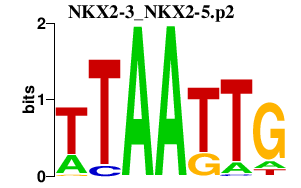
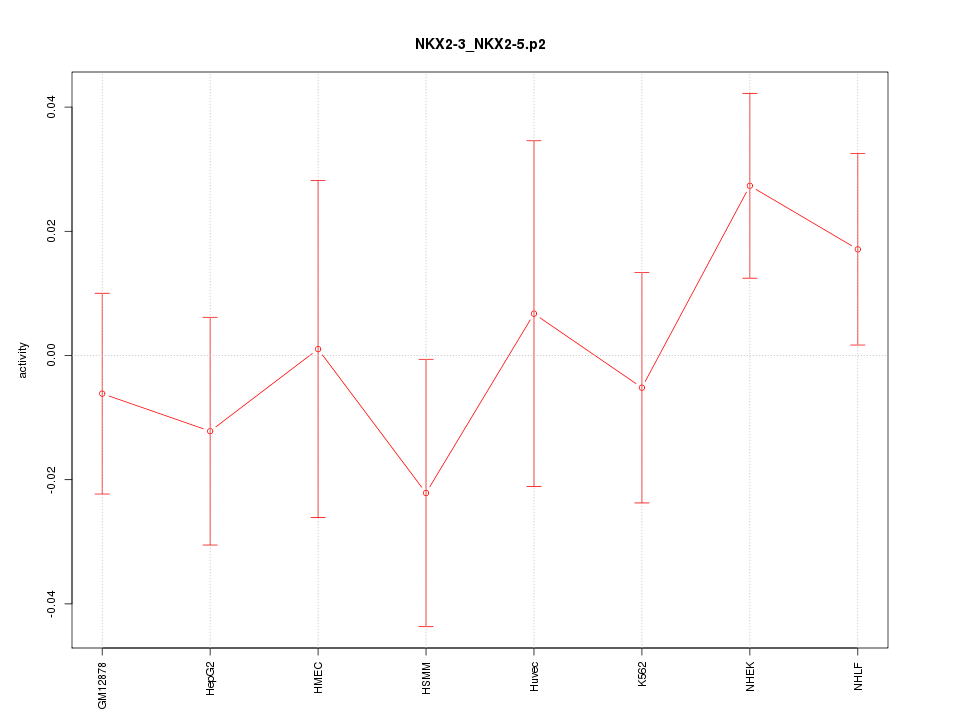
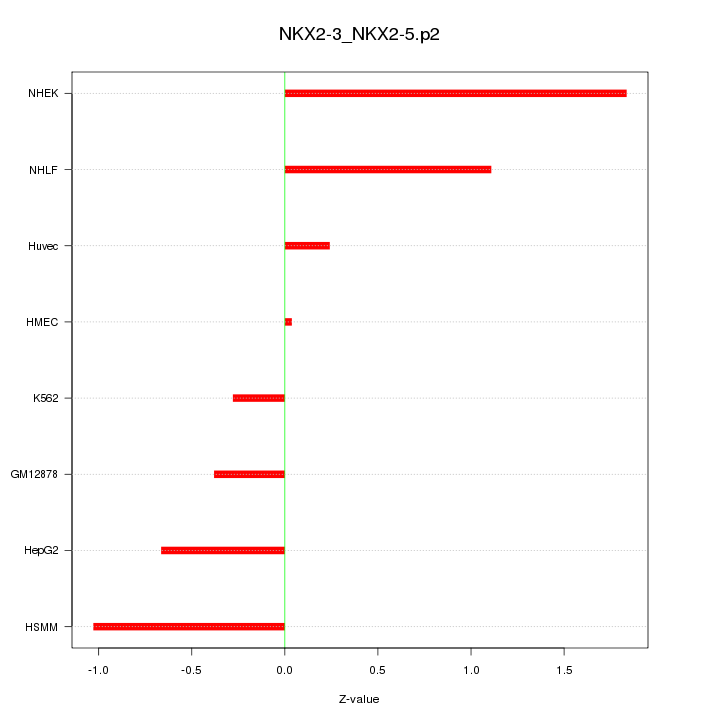
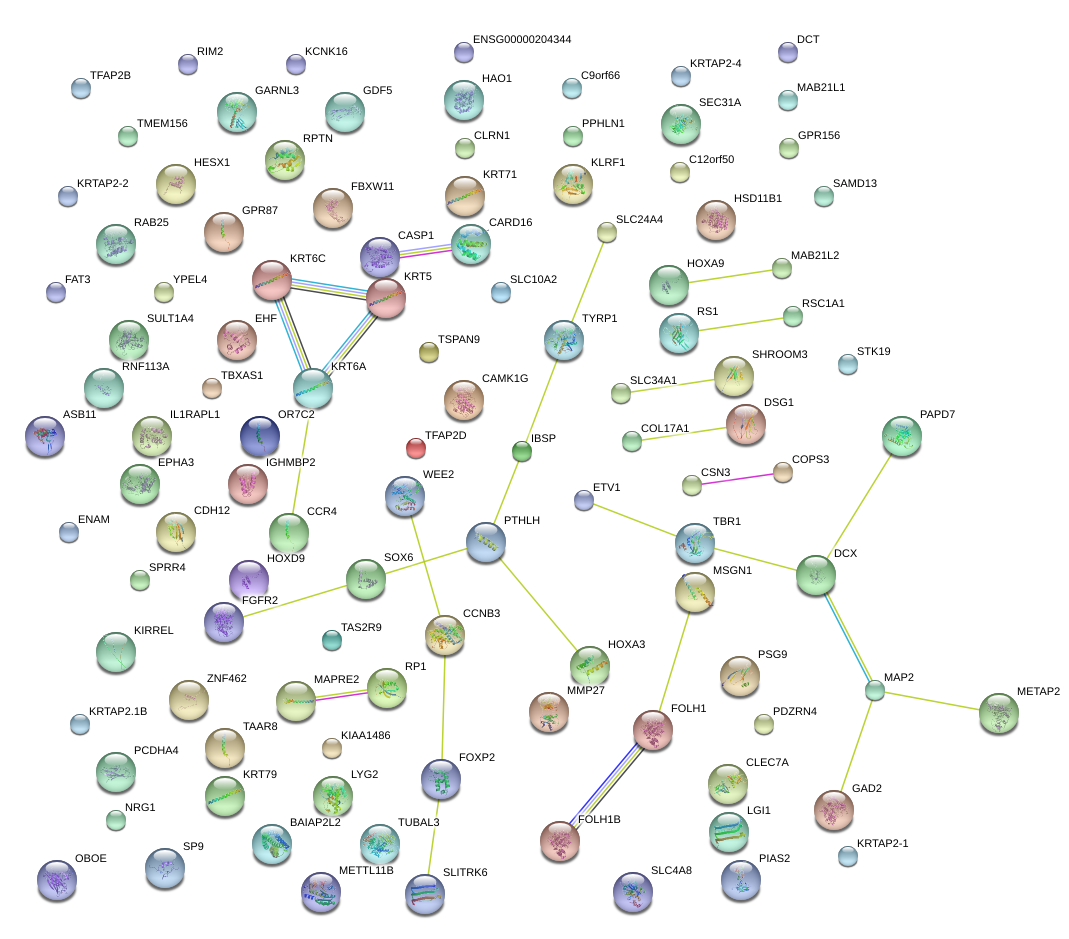
|
chr3_-_152517204
|
1.403
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr7_-_13992351
|
1.312
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr7_-_27171673
|
1.215
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171648
|
1.186
|
|
HOXA9
|
homeobox A9
|
|
chr3_+_89239494
|
1.150
|
|
EPHA3
|
EPH receptor A3
|
|
chr18_+_27152049
|
1.090
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr13_-_102517196
|
1.081
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr3_-_57209319
|
1.056
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr4_+_71713324
|
1.010
|
NM_031889
|
ENAM
|
enamelin
|
|
chr12_-_51173238
|
1.010
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chrX_-_110541029
|
0.995
|
NM_000555
|
DCX
|
doublecortin
|
|
chr10_-_105835525
|
0.969
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr8_+_104900591
|
0.888
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr9_+_108726257
|
0.875
|
|
ZNF462
|
zinc finger protein 462
|
|
chrX_+_28515479
|
0.829
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr4_-_38710376
|
0.805
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr6_+_50789215
|
0.804
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chrX_-_15243647
|
0.786
|
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr2_+_174907914
|
0.763
|
NM_001145250
|
SP9
|
Sp9 transcription factor homolog (mouse)
|
|
chr2_+_210152609
|
0.762
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr11_-_102081654
|
0.758
|
NM_022122
|
MMP27
|
matrix metallopeptidase 27
|
|
chr3_-_121445831
|
0.745
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr11_+_91724909
|
0.732
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr12_-_86947276
|
0.720
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr7_-_27171600
|
0.718
|
|
HOXA9
|
homeobox A9
|
|
chr6_+_134800533
|
0.713
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr10_-_105835609
|
0.711
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr13_-_85271329
|
0.705
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr6_-_39398293
|
0.681
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr13_-_93929719
|
0.671
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr9_+_129066576
|
0.639
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr6_+_50894397
|
0.638
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr1_+_156229686
|
0.630
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr1_+_151209751
|
0.619
|
NM_173080
|
SPRR4
|
small proline-rich protein 4
|
|
chr2_+_225973845
|
0.612
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr12_-_28016182
|
0.611
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr5_-_168204430
|
0.609
|
|
|
|
|
chr5_+_140166842
|
0.601
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr13_-_34948758
|
0.597
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr12_+_9871343
|
0.587
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr7_+_113842284
|
0.581
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chrX_-_18600108
|
0.573
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr6_-_25679197
|
0.571
|
|
|
|
|
chr9_+_12683385
|
0.569
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr10_+_26545241
|
0.564
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr2_+_161980865
|
0.556
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_27133163
|
0.550
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr6_+_132915522
|
0.549
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr12_-_51200347
|
0.546
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr12_-_51233001
|
0.546
|
NM_033448
|
KRT71
|
keratin 71
|
|
chr12_+_40117751
|
0.537
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr3_-_152173475
|
0.532
|
NM_001195794
NM_174878
|
CLRN1
|
clarin 1
|
|
chr1_+_207823667
|
0.527
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr15_-_64108718
|
0.526
|
|
|
|
|
chr1_+_207944807
|
0.526
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr2_+_161981138
|
0.524
|
|
TBR1
|
T-box, brain, 1
|
|
chr1_+_154297588
|
0.521
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr10_-_5436781
|
0.517
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr10_+_95507555
|
0.516
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr14_+_91858677
|
0.513
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_+_89239363
|
0.510
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr9_-_205892
|
0.509
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr17_-_36456949
|
0.507
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr11_+_34599227
|
0.505
|
|
EHF
|
ets homologous factor
|
|
chr11_+_89032112
|
0.503
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr4_+_77575276
|
0.501
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr5_+_176744037
|
0.499
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr10_-_123346148
|
0.495
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr20_-_33489383
|
0.492
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr4_+_71142893
|
0.485
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr1_+_84540921
|
0.480
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chrX_+_50044279
|
0.475
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr2_+_17861266
|
0.473
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr11_-_104411065
|
0.471
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr12_-_10173947
|
0.470
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr11_-_16454493
|
0.468
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_57173823
|
0.467
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr1_-_150398327
|
0.466
|
NM_001122965
|
RPTN
|
repetin
|
|
chr9_+_108665168
|
0.461
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr12_-_10853940
|
0.460
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr5_-_22889487
|
0.458
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr7_+_141054621
|
0.456
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr7_+_139175420
|
0.455
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr1_+_168381811
|
0.453
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr22_-_36836620
|
0.452
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr20_-_7869068
|
0.451
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr4_+_88939725
|
0.448
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr2_-_99238001
|
0.445
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr8_+_55691179
|
0.445
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr4_+_68995761
|
0.444
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr3_+_14835472
|
0.442
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr1_-_155336223
|
0.438
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr4_+_94969064
|
0.434
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr13_-_45323757
|
0.434
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr13_+_77007809
|
0.434
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr15_-_96875133
|
0.431
|
NM_182562
|
FAM169B
|
family with sequence similarity 169, member B
|
|
chr3_+_162697289
|
0.427
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr5_+_31229609
|
0.426
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr17_-_36877002
|
0.425
|
NM_002278
|
KRT32
|
keratin 32
|
|
chrX_+_153101278
|
0.420
|
NM_000513
NM_001048181
|
OPN1MW
OPN1MW2
|
opsin 1 (cone pigments), medium-wave-sensitive
opsin 1 (cone pigments), medium-wave-sensitive 2
|
|
chr12_+_8866416
|
0.419
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr13_-_37070862
|
0.419
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr7_-_115458010
|
0.418
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr2_+_234490781
|
0.416
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr8_-_40874485
|
0.415
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr12_-_6103945
|
0.409
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr8_-_102872354
|
0.409
|
|
NCALD
|
neurocalcin delta
|
|
chr12_-_51086330
|
0.409
|
NM_033033
|
KRT82
|
keratin 82
|
|
chr7_-_36991189
|
0.408
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr12_-_16650687
|
0.404
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_13996166
|
0.403
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr5_+_140583098
|
0.395
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_+_140703784
|
0.391
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr11_-_46364681
|
0.386
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr5_-_22248630
|
0.386
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr4_+_9094611
|
0.385
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr1_+_117098579
|
0.384
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr9_+_124173049
|
0.383
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr4_+_30331289
|
0.381
|
|
PCDH7
|
protocadherin 7
|
|
chr6_+_136214495
|
0.376
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr3_+_85858321
|
0.376
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr17_-_64649608
|
0.374
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr13_+_52500830
|
0.365
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr3_+_190990142
|
0.365
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chrX_-_106033202
|
0.360
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr5_+_140848905
|
0.359
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr21_-_32879617
|
0.359
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr15_+_92700167
|
0.358
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr15_+_56217686
|
0.356
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr10_-_21475493
|
0.355
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr10_+_98054074
|
0.352
|
NM_001017520
NM_004088
|
DNTT
|
deoxynucleotidyltransferase, terminal
|
|
chr1_-_154665807
|
0.350
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr13_-_60887655
|
0.350
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr4_+_75393053
|
0.349
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chrX_+_90976314
|
0.349
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr5_-_39255335
|
0.347
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr1_-_224993498
|
0.346
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr2_-_207291364
|
0.345
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr10_-_50066386
|
0.342
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr2_+_234266250
|
0.342
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr19_+_7874701
|
0.340
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr1_+_145841569
|
0.338
|
NM_005267
|
GJA8
|
gap junction protein, alpha 8, 50kDa
|
|
chr11_+_128067598
|
0.336
|
NM_001167681
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr21_-_31175722
|
0.334
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr17_-_36347197
|
0.331
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_-_123353607
|
0.330
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr1_+_151240846
|
0.328
|
NM_001097589
NM_005416
|
SPRR3
|
small proline-rich protein 3
|
|
chr20_+_62266217
|
0.327
|
|
MYT1
|
myelin transcription factor 1
|
|
chr1_+_74473672
|
0.326
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr17_-_76064842
|
0.324
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr11_-_56166490
|
0.324
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr3_-_74652952
|
0.323
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr17_-_39191658
|
0.322
|
NM_025237
|
SOST
|
sclerostin
|
|
chr8_-_20084892
|
0.322
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr17_-_19290745
|
0.321
|
|
|
|
|
chr3_+_46370578
|
0.321
|
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr6_-_136888771
|
0.320
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr2_+_161981046
|
0.319
|
|
TBR1
|
T-box, brain, 1
|
|
chr6_-_155818728
|
0.319
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr14_+_91859289
|
0.319
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr10_+_102212787
|
0.319
|
NM_003393
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr4_-_70547962
|
0.316
|
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr16_-_4792857
|
0.315
|
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr19_-_3723208
|
0.314
|
NM_032753
|
RAX2
|
retina and anterior neural fold homeobox 2
|
|
chr5_-_1935764
|
0.313
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr9_-_70345549
|
0.312
|
NM_153237
|
C9orf71
|
chromosome 9 open reading frame 71
|
|
chr13_+_45174446
|
0.312
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr6_+_29187565
|
0.310
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr7_-_97457286
|
0.310
|
NM_006188
|
OCM2
|
oncomodulin 2
|
|
chr11_+_4799126
|
0.310
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr13_-_19703371
|
0.310
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr16_+_65938753
|
0.308
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr14_-_56342097
|
0.308
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr15_-_48345453
|
0.307
|
NM_002112
|
HDC
|
histidine decarboxylase
|
|
chr14_-_56346849
|
0.306
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chrX_+_30158473
|
0.305
|
NM_002365
|
MAGEB3
|
melanoma antigen family B, 3
|
|
chr2_+_210152399
|
0.305
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_+_140187746
|
0.303
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr2_-_73373956
|
0.301
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr7_-_31347032
|
0.300
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr11_-_83071084
|
0.299
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr17_+_71232370
|
0.298
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chrX_-_128616594
|
0.298
|
NM_017413
|
APLN
|
apelin
|
|
chr8_-_72436832
|
0.295
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_+_166034402
|
0.294
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chrX_+_102852492
|
0.294
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr21_+_39117064
|
0.293
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr3_-_188492163
|
0.292
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr14_-_100105883
|
0.289
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr5_-_149649380
|
0.288
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr3_+_160269836
|
0.288
|
|
IQCJ
|
IQ motif containing J
|
|
chr11_+_34599163
|
0.285
|
NM_012153
|
EHF
|
ets homologous factor
|
|
chr17_+_56843893
|
0.285
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr7_-_41709191
|
0.285
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr14_+_19473606
|
0.285
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr11_-_16380967
|
0.282
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_-_48212683
|
0.280
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr5_-_149649593
|
0.279
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr11_+_88550687
|
0.279
|
NM_000372
|
TYR
|
tyrosinase (oculocutaneous albinism IA)
|
|
chr14_-_36121536
|
0.276
|
NM_014360
|
NKX2-8
|
NK2 homeobox 8
|