|
chr4_+_169654742
|
2.057
|
NM_001166108
NM_016081
|
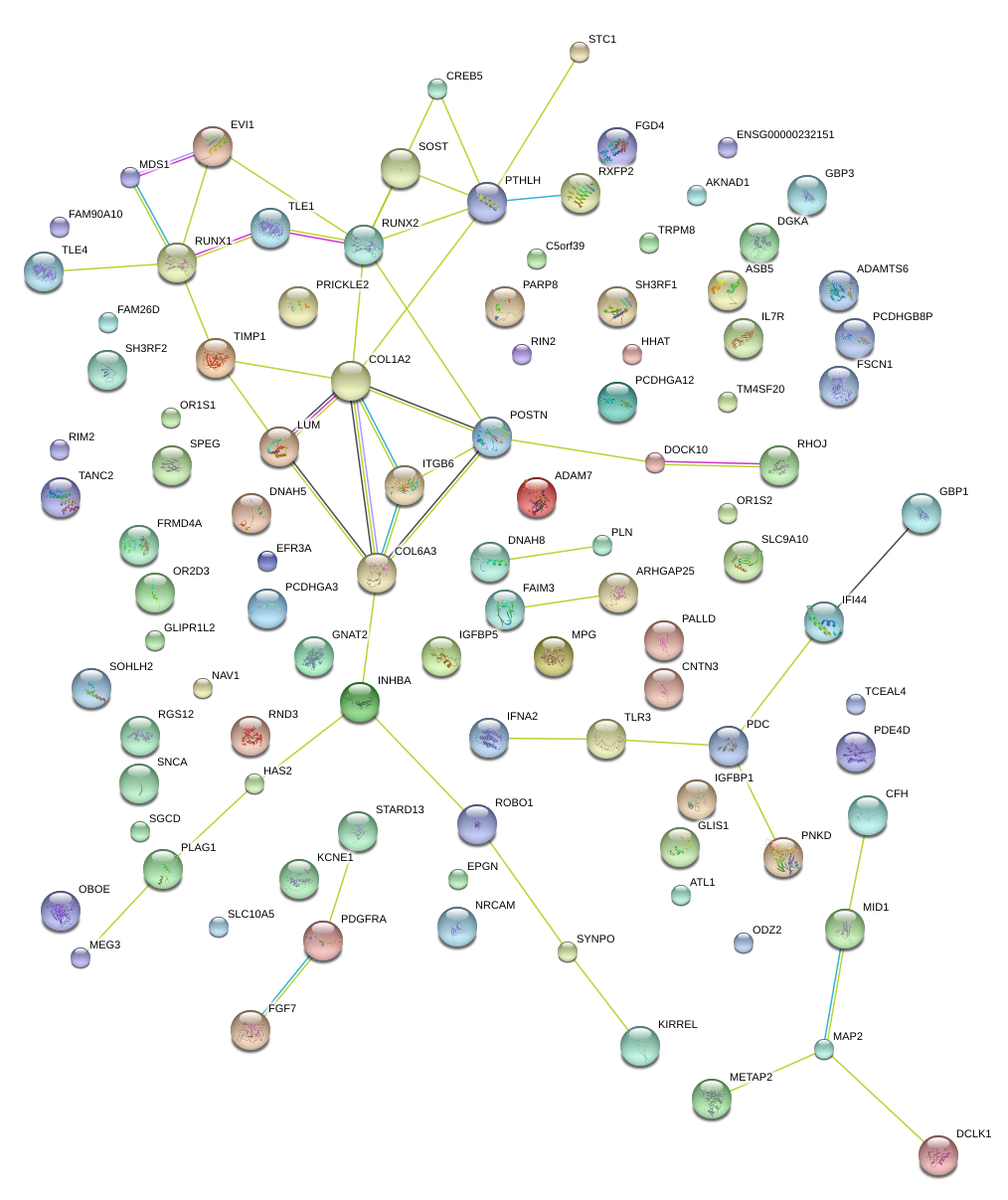
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr2_-_160764819
|
1.917
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr2_-_160765008
|
1.384
|
|
ITGB6
|
integrin, beta 6
|
|
chr5_+_166644420
|
1.267
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr3_-_64186151
|
1.233
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_-_151052391
|
1.153
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr7_-_41709191
|
1.147
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr4_-_90978469
|
1.047
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chrX_-_10495476
|
1.033
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_237987579
|
0.907
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr12_+_32546243
|
0.899
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr17_+_58440629
|
0.860
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr2_-_237987554
|
0.853
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr20_+_19863716
|
0.831
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr8_-_122722674
|
0.827
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr5_-_58331515
|
0.778
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_177427252
|
0.712
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr2_-_237987545
|
0.710
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_+_58625086
|
0.700
|
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr7_+_28692122
|
0.693
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_+_81376666
|
0.680
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chrX_-_10504852
|
0.640
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_-_28016182
|
0.621
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr14_+_100365390
|
0.612
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_+_199884152
|
0.600
|
|
NAV1
|
neuron navigator 1
|
|
chr9_-_21375258
|
0.600
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr5_+_149977398
|
0.597
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr5_-_64813459
|
0.586
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr7_-_107667728
|
0.586
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_+_54790190
|
0.573
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr7_+_28305464
|
0.571
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_+_133060353
|
0.569
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chrX_+_102727068
|
0.546
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr14_+_100365162
|
0.546
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr4_+_75393053
|
0.537
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr15_+_47502666
|
0.536
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr1_+_208568218
|
0.517
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr1_-_89261021
|
0.512
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr10_-_14412813
|
0.508
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_+_93862137
|
0.507
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr3_-_74652952
|
0.505
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr8_-_23768242
|
0.502
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr2_-_237987553
|
0.496
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_-_39191658
|
0.494
|
NM_025237
|
SOST
|
sclerostin
|
|
chr21_-_35343448
|
0.481
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr8_-_82769761
|
0.477
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr21_-_35343456
|
0.467
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr5_+_145296313
|
0.463
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr6_+_118976134
|
0.459
|
NM_002667
|
PLN
|
phospholamban
|
|
chr7_+_5609416
|
0.456
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr13_-_32658215
|
0.455
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_+_56406296
|
0.448
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr21_-_35343500
|
0.443
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr12_+_54611212
|
0.440
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr3_-_79899748
|
0.434
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr13_-_37070862
|
0.434
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr1_-_109957195
|
0.431
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr2_-_227952249
|
0.429
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr12_+_74071116
|
0.428
|
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr1_-_109201228
|
0.427
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr1_+_78888064
|
0.423
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr5_-_43075904
|
0.420
|
NM_001014279
|
C5orf39
|
chromosome 5 open reading frame 39
|
|
chr8_+_104900591
|
0.417
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr5_+_49997489
|
0.417
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_156229686
|
0.411
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr3_-_113495763
|
0.407
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr6_+_116956868
|
0.406
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr14_+_62740878
|
0.405
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr2_-_217268492
|
0.405
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr5_-_13997588
|
0.405
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr5_+_35892747
|
0.397
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr12_-_90029328
|
0.389
|
|
LUM
|
lumican
|
|
chr1_+_194887630
|
0.389
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr2_-_225519974
|
0.388
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr5_+_140780797
|
0.364
|
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr2_-_217267742
|
0.364
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chrX_+_47326746
|
0.363
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr13_-_35327798
|
0.361
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr1_-_205161821
|
0.354
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr21_-_34806373
|
0.349
|
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr11_+_6898780
|
0.342
|
NM_001004684
|
OR2D3
|
olfactory receptor, family 2, subfamily D, member 3
|
|
chr2_+_68815416
|
0.341
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr5_+_155686333
|
0.341
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr13_-_35686665
|
0.341
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chr13_+_31211678
|
0.340
|
NM_001166058
NM_130806
|
RXFP2
|
relaxin/insulin-like family peptide receptor 2
|
|
chr2_+_220017617
|
0.336
|
|
SPEG
|
SPEG complex locus
|
|
chr7_+_45899080
|
0.336
|
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr4_-_170314382
|
0.335
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr3_-_170348215
|
0.335
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr4_+_3285671
|
0.333
|
NM_002926
NM_198229
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr5_+_140719773
|
0.332
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr11_-_57728228
|
0.331
|
NM_001004459
|
OR1S2
|
olfactory receptor, family 1, subfamily S, member 2
|
|
chr1_-_53972464
|
0.330
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr5_+_140762551
|
0.329
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr8_+_24354387
|
0.329
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr4_+_187227302
|
0.324
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr2_+_234490781
|
0.323
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr4_+_54790020
|
0.322
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr2_+_210152609
|
0.318
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_-_184696839
|
0.317
|
NM_002597
|
PDC
|
phosducin
|
|
chr12_-_10464308
|
0.315
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr12_-_90029672
|
0.315
|
NM_002345
|
LUM
|
lumican
|
|
chr9_+_27099138
|
0.314
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chrX_+_47326594
|
0.313
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr3_+_150065732
|
0.311
|
NM_001870
|
CPA3
|
carboxypeptidase A3 (mast cell)
|
|
chr7_+_128099581
|
0.309
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr10_-_90601655
|
0.304
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr18_+_59705918
|
0.301
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr10_+_64894943
|
0.301
|
|
LOC84989
|
hypothetical LOC84989
|
|
chr21_+_29439768
|
0.299
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr6_+_125346208
|
0.295
|
NM_152553
|
RNF217
|
ring finger protein 217
|
|
chr4_-_87593306
|
0.288
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr6_+_88026338
|
0.287
|
|
ZNF292
|
zinc finger protein 292
|
|
chr17_-_10217046
|
0.286
|
NM_003802
|
MYH13
|
myosin, heavy chain 13, skeletal muscle
|
|
chr12_-_16652202
|
0.286
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_36559579
|
0.282
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr11_+_122258600
|
0.276
|
NM_024806
NM_199124
|
C11orf63
|
chromosome 11 open reading frame 63
|
|
chr13_-_109600709
|
0.275
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr8_+_79591074
|
0.275
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr5_+_140546839
|
0.275
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr13_+_99027366
|
0.273
|
|
|
|
|
chr5_+_140848905
|
0.273
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr19_-_47623340
|
0.270
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr8_+_11225904
|
0.268
|
NM_054028
|
AMAC1L2
|
acyl-malonyl condensing enzyme 1-like 2
|
|
chr1_-_67366807
|
0.267
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr6_-_31242593
|
0.266
|
NM_001173531
NM_203289
|
POU5F1
|
POU class 5 homeobox 1
|
|
chr15_+_78520634
|
0.265
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr18_-_49944986
|
0.263
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr1_+_203179976
|
0.261
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr3_-_148607096
|
0.260
|
NM_032153
|
ZIC4
|
Zic family member 4
|
|
chr11_-_128399218
|
0.260
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr11_-_27637771
|
0.259
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_-_120413798
|
0.258
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr5_+_140780720
|
0.257
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr4_-_77147625
|
0.256
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr4_+_120167721
|
0.255
|
|
SYNPO2
|
synaptopodin 2
|
|
chr7_+_120752656
|
0.255
|
NM_016087
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr3_+_85858321
|
0.255
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr1_+_207668787
|
0.252
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr1_+_170656450
|
0.252
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr9_+_81377289
|
0.251
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr2_+_135312655
|
0.251
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chrX_-_114158388
|
0.251
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr3_-_121445831
|
0.248
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr4_-_138673078
|
0.246
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_219982081
|
0.244
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_21249974
|
0.243
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr13_-_46368968
|
0.243
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr1_-_219982016
|
0.242
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr11_+_55629094
|
0.242
|
NM_001005200
|
OR8H2
|
olfactory receptor, family 8, subfamily H, member 2
|
|
chr8_+_67507271
|
0.241
|
NM_144650
|
ADHFE1
|
alcohol dehydrogenase, iron containing, 1
|
|
chr17_-_36376663
|
0.240
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr3_+_133518900
|
0.239
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr2_-_162808123
|
0.237
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr8_+_42671718
|
0.237
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr10_-_96819064
|
0.237
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chrX_+_135058402
|
0.236
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr8_-_131483395
|
0.236
|
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr20_+_9146036
|
0.235
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr5_+_140790310
|
0.231
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr8_+_79590890
|
0.231
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr8_-_72431274
|
0.229
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr15_-_74091817
|
0.228
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr6_-_37773609
|
0.227
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chrX_-_119550402
|
0.227
|
|
CUL4B
|
cullin 4B
|
|
chr3_-_21767819
|
0.226
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr1_+_78858675
|
0.225
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr11_-_7941617
|
0.223
|
NM_176821
|
NLRP10
|
NLR family, pyrin domain containing 10
|
|
chr6_-_152744143
|
0.223
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_+_180685878
|
0.222
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr17_-_7248137
|
0.222
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 read-through transcript
chromosome 17 open reading frame 61
|
|
chr2_+_61243132
|
0.220
|
NM_001143959
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr6_+_114285209
|
0.220
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chrX_-_19898302
|
0.219
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr10_-_69422391
|
0.218
|
|
HERC4
|
hect domain and RLD 4
|
|
chr4_-_70760381
|
0.214
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr6_+_12398514
|
0.213
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr2_+_17861266
|
0.213
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr14_-_70071484
|
0.212
|
NM_003814
|
ADAM20
|
ADAM metallopeptidase domain 20
|
|
chr3_+_190990142
|
0.212
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr7_-_22828922
|
0.210
|
NM_019059
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast)
|
|
chr7_-_7541974
|
0.210
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr4_-_159312973
|
0.206
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_189263395
|
0.206
|
NM_173553
|
TRIML2
|
tripartite motif family-like 2
|
|
chr4_-_100492854
|
0.206
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr18_-_45272820
|
0.205
|
|
RPL17
|
ribosomal protein L17
|
|
chrX_-_10605726
|
0.204
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr19_+_24061717
|
0.203
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr2_-_202271598
|
0.203
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr11_-_101906640
|
0.202
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr1_+_205068844
|
0.202
|
NM_013371
|
IL19
|
interleukin 19
|
|
chr1_-_32054164
|
0.202
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chrX_+_80343958
|
0.202
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr3_-_116272911
|
0.200
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_111253484
|
0.200
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr1_+_33319287
|
0.199
|
NM_052998
|
ADC
|
arginine decarboxylase
|
|
chr12_+_27288344
|
0.196
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr5_+_76418214
|
0.196
|
|
LOC728723
|
hypothetical LOC728723
|
|
chr3_+_149930656
|
0.194
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr4_-_77097582
|
0.192
|
|
SDAD1
|
SDA1 domain containing 1
|