|
chr7_+_5289079
|
1.689
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr21_+_46226072
|
1.490
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr19_+_56506907
|
1.385
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr20_-_30535045
|
1.365
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr6_+_37245860
|
1.314
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr2_+_42128521
|
1.313
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr20_-_30534848
|
1.300
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr5_-_1165106
|
1.280
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr13_-_113066366
|
1.090
|
NM_024719
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr18_-_24011349
|
1.087
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr14_-_20636568
|
1.083
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr20_-_62181588
|
1.068
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr19_-_48835783
|
1.066
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr3_+_185580554
|
1.032
|
NM_003741
|
CHRD
|
chordin
|
|
chr16_-_70475381
|
1.029
|
|
ZNF821
|
zinc finger protein 821
|
|
chr19_-_59676153
|
1.015
|
NM_145057
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr12_+_3056781
|
0.981
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr19_+_50041424
|
0.980
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr1_-_158335241
|
0.977
|
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr9_+_130223046
|
0.970
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr18_-_24010944
|
0.953
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chrX_-_1291526
|
0.950
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr7_+_72720093
|
0.948
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr1_-_158334983
|
0.945
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chrX_+_9392980
|
0.936
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_-_3102169
|
0.933
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr10_+_114699953
|
0.904
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr12_+_55808514
|
0.902
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr14_-_20636337
|
0.879
|
|
ZNF219
|
zinc finger protein 219
|
|
chr19_+_50041232
|
0.878
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr19_+_59064471
|
0.878
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr20_-_62181231
|
0.866
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr22_+_49459935
|
0.865
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chrX_+_152643516
|
0.856
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr7_+_72720256
|
0.853
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr19_+_47040863
|
0.850
|
NM_001040283
|
DMRTC2
|
DMRT-like family C2
|
|
chr19_+_40213431
|
0.826
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr9_-_125732005
|
0.825
|
NM_020946
NM_024820
|
DENND1A
|
DENN/MADD domain containing 1A
|
|
chr19_-_55524445
|
0.813
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chrX_+_138060
|
0.810
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr1_-_11637325
|
0.809
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr17_+_52688842
|
0.805
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr16_-_88613287
|
0.804
|
NM_001042610
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr8_-_144722141
|
0.796
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr4_+_1764802
|
0.792
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr19_+_59064795
|
0.791
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_-_23623713
|
0.789
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr1_-_11637058
|
0.785
|
|
FBXO2
|
F-box protein 2
|
|
chr20_+_3661316
|
0.769
|
NM_001197327
NM_052970
|
HSPA12B
|
heat shock 70kD protein 12B
|
|
chr16_+_88542639
|
0.768
|
NM_017702
NM_207514
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr19_+_59062962
|
0.763
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_-_158334885
|
0.762
|
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr17_-_76622967
|
0.758
|
|
FLJ90757
|
hypothetical LOC440465
|
|
chr4_+_3264509
|
0.757
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr7_-_51351825
|
0.755
|
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr7_-_100077072
|
0.751
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr12_-_47468985
|
0.749
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr19_+_61344501
|
0.743
|
|
ZNF444
|
zinc finger protein 444
|
|
chr9_+_130222943
|
0.735
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr20_+_34357604
|
0.723
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr7_-_158073121
|
0.717
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr19_+_38979502
|
0.711
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr15_+_78232194
|
0.710
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr11_+_65443303
|
0.709
|
NM_006442
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr19_+_61344341
|
0.708
|
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr17_+_34114924
|
0.708
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr16_-_30014993
|
0.703
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr22_-_19122107
|
0.702
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr19_-_1518875
|
0.699
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr22_-_19122047
|
0.689
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr15_+_78232232
|
0.686
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr22_+_48951286
|
0.684
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr11_+_56984702
|
0.682
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr1_-_11636983
|
0.674
|
|
FBXO2
|
F-box protein 2
|
|
chr14_+_20608179
|
0.665
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr14_-_37134191
|
0.663
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr12_+_6800950
|
0.662
|
NM_014449
NM_019858
|
GPR162
|
G protein-coupled receptor 162
|
|
chr19_+_18069596
|
0.662
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr3_-_52419086
|
0.661
|
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr17_-_73636362
|
0.660
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr5_+_172193826
|
0.657
|
NM_001031711
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr9_+_137511448
|
0.657
|
NM_014811
|
KIAA0649
|
KIAA0649
|
|
chr2_-_218551786
|
0.653
|
|
TNS1
|
tensin 1
|
|
chr19_+_59062937
|
0.652
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr17_+_56832255
|
0.644
|
|
TBX2
|
T-box 2
|
|
chr11_-_74914514
|
0.641
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chrX_+_2756810
|
0.634
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr20_-_51644187
|
0.631
|
|
ZNF217
|
zinc finger protein 217
|
|
chr17_+_34115383
|
0.630
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr12_+_55896844
|
0.626
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr3_-_14964403
|
0.625
|
|
|
|
|
chr11_+_359723
|
0.624
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr16_-_11257539
|
0.621
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr17_-_75385371
|
0.618
|
NM_020649
|
CBX8
|
chromobox homolog 8
|
|
chr11_-_60819110
|
0.617
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr16_-_51138306
|
0.616
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr2_-_200030944
|
0.615
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr19_-_52426055
|
0.608
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr15_+_78232160
|
0.607
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr14_+_104852225
|
0.607
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr2_+_218896219
|
0.605
|
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr7_+_6622051
|
0.603
|
NM_017560
|
ZNF853
|
zinc finger protein 853
|
|
chr12_+_2774550
|
0.601
|
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr19_+_52451642
|
0.600
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr5_+_109052945
|
0.598
|
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr17_-_39632845
|
0.597
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr11_-_60819273
|
0.597
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr1_-_21868380
|
0.596
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_+_7891884
|
0.595
|
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa
|
|
chr7_-_103417198
|
0.595
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr14_-_21064182
|
0.590
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr9_-_129370954
|
0.588
|
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr20_+_4077409
|
0.583
|
NM_175839
NM_175840
NM_175841
NM_175842
|
SMOX
|
spermine oxidase
|
|
chr10_-_95350949
|
0.578
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr20_+_4077466
|
0.574
|
|
SMOX
|
spermine oxidase
|
|
chr1_+_243200251
|
0.570
|
NM_001143943
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr2_+_63131466
|
0.569
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr20_+_60906605
|
0.568
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr19_-_18578631
|
0.567
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr17_+_44429748
|
0.567
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr16_+_49139694
|
0.561
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr6_+_44203335
|
0.559
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr16_-_49742651
|
0.558
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr16_+_1299180
|
0.555
|
NM_194261
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr19_-_18578577
|
0.553
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr16_+_87741416
|
0.551
|
|
|
|
|
chr6_-_44203155
|
0.549
|
NM_032111
|
MRPL14
|
mitochondrial ribosomal protein L14
|
|
chr17_-_7061641
|
0.545
|
NM_001128827
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr2_-_166940710
|
0.545
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr17_-_34157962
|
0.543
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr20_+_30813851
|
0.542
|
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr2_-_42574587
|
0.541
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr6_-_41855564
|
0.533
|
NM_006653
|
FRS3
|
fibroblast growth factor receptor substrate 3
|
|
chr19_+_54708389
|
0.533
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr16_+_87047159
|
0.531
|
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr3_+_50279993
|
0.531
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr15_-_43267441
|
0.530
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr12_-_123023312
|
0.527
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr16_-_29817841
|
0.526
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr16_+_29892714
|
0.523
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr16_+_88422354
|
0.522
|
NM_032451
|
SPIRE2
|
spire homolog 2 (Drosophila)
|
|
chr2_+_218896102
|
0.521
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr20_+_29526751
|
0.520
|
NM_014012
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1
|
|
chr19_+_40223249
|
0.518
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr1_+_224317036
|
0.516
|
NM_002107
|
H3F3A
|
H3 histone, family 3A
|
|
chr16_+_1299361
|
0.514
|
NM_194260
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr12_+_55808696
|
0.513
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr20_-_22512893
|
0.512
|
|
FOXA2
|
forkhead box A2
|
|
chr1_-_44912704
|
0.511
|
|
TMEM53
|
transmembrane protein 53
|
|
chr14_-_37134050
|
0.510
|
|
FOXA1
|
forkhead box A1
|
|
chr11_-_64247230
|
0.510
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr3_+_185536376
|
0.510
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr11_-_70641267
|
0.509
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr19_+_47480445
|
0.508
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr19_-_41215380
|
0.508
|
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr9_+_130222511
|
0.506
|
NM_016174
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr17_+_7252225
|
0.506
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr17_-_38978253
|
0.506
|
|
ETV4
|
ets variant 4
|
|
chr22_+_47350781
|
0.505
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr16_+_29730909
|
0.505
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr3_+_13496714
|
0.504
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr19_+_52451570
|
0.503
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr2_-_239987629
|
0.503
|
|
HDAC4
|
histone deacetylase 4
|
|
chr9_+_136358145
|
0.502
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr22_+_49293520
|
0.502
|
|
NCAPH2
|
non-SMC condensin II complex, subunit H2
|
|
chr19_-_14062230
|
0.500
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr20_+_62181878
|
0.500
|
NM_000913
NM_182647
|
OPRL1
|
opiate receptor-like 1
|
|
chr19_+_12995782
|
0.499
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr22_-_30072329
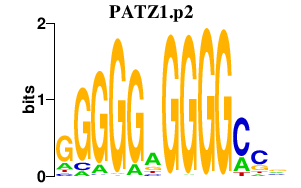
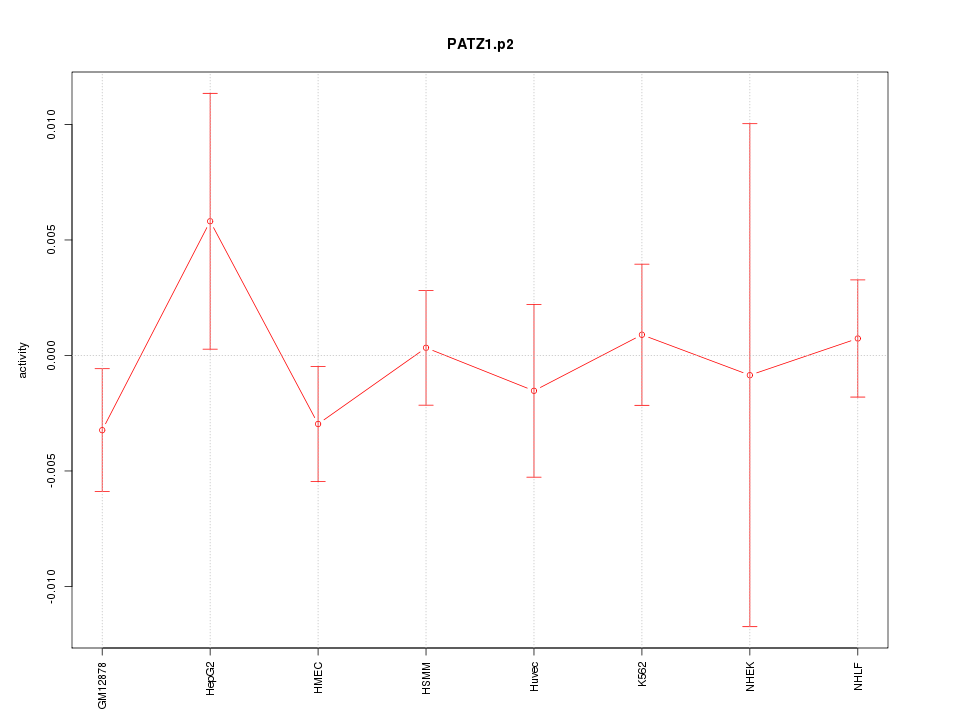
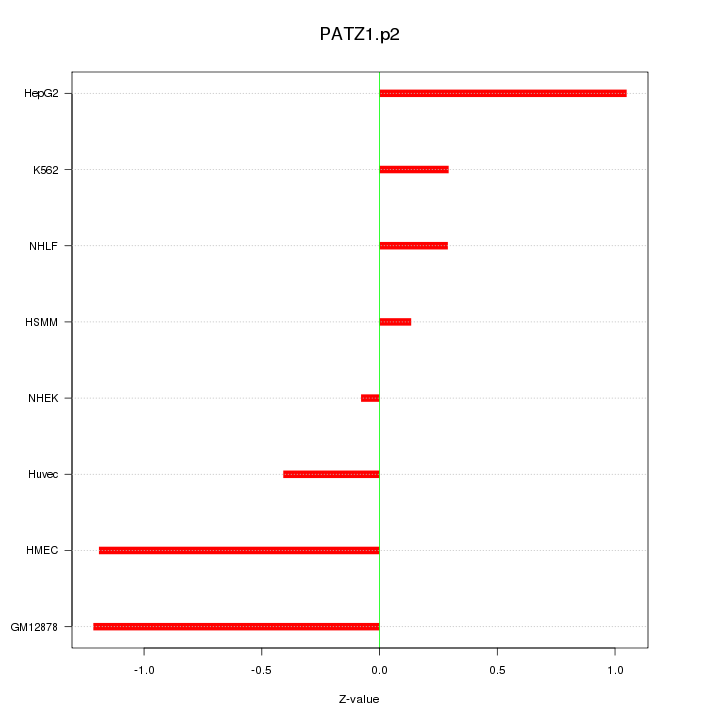
|
0.498
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr1_-_32574185
|
0.496
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr19_+_40958206
|
0.496
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr20_+_30814048
|
0.495
|
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr7_+_73080362
|
0.495
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr2_+_70981233
|
0.495
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr19_+_11327061
|
0.490
|
NM_001170635
NM_022737
|
LPPR2
|
lipid phosphate phosphatase-related protein type 2
|
|
chr2_-_216008689
|
0.490
|
|
FN1
|
fibronectin 1
|
|
chr11_-_86343859
|
0.489
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr7_+_100156358
|
0.489
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr1_+_160306093
|
0.488
|
NM_001164757
NM_014697
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr20_-_17610704
|
0.486
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr15_-_43267139
|
0.486
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr9_+_132961723
|
0.484
|
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr3_-_14964620
|
0.483
|
|
|
|
|
chr10_+_134201262
|
0.482
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr7_-_74705276
|
0.481
|
NM_001145064
|
GATSL2
|
GATS protein-like 2
|
|
chr17_-_78249750
|
0.480
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr19_+_59062982
|
0.479
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr8_+_145700272
|
0.479
|
NM_005309
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase)
|
|
chr6_+_7053183
|
0.478
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr16_+_31036474
|
0.478
|
NM_032188
NM_182958
|
MYST1
|
MYST histone acetyltransferase 1
|
|
chr12_+_56621711
|
0.475
|
NM_033276
|
XRCC6BP1
|
XRCC6 binding protein 1
|
|
chr5_+_176446425
|
0.474
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr18_+_44319365
|
0.474
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr6_-_44203131
|
0.474
|
|
MRPL14
|
mitochondrial ribosomal protein L14
|
|
chr17_-_40401138
|
0.472
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr2_+_112372671
|
0.472
|
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr14_-_20636909
|
0.469
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr1_-_54644663
|
0.469
|
NM_001009955
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr17_-_63965156
|
0.468
|
NM_017983
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr17_-_38978772
|
0.467
|
NM_001986
|
ETV4
|
ets variant 4
|