|
chr11_-_85457469
|
1.520
|
|
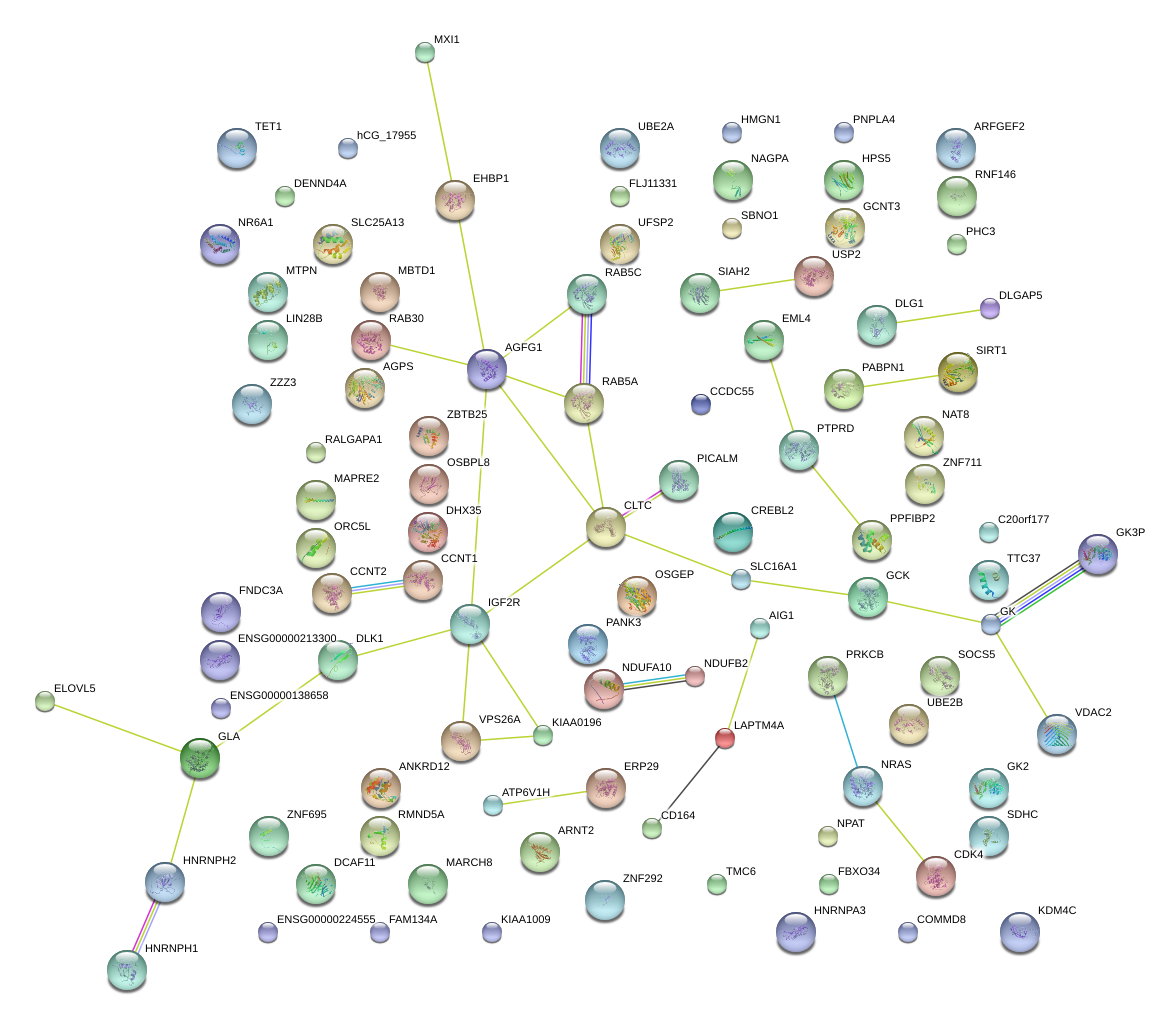
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr5_+_133734768
|
1.474
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr6_+_160310118
|
1.316
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr20_+_46971618
|
1.288
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr11_+_7574992
|
1.284
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr11_-_85457753
|
1.281
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr11_-_85457736
|
1.243
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_56432385
|
1.210
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr2_+_46779553
|
1.195
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr14_+_23654161
|
1.149
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr15_-_63871492
|
1.143
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr17_+_55051818
|
1.128
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr15_-_63871598
|
1.079
|
NM_001144823
NM_005848
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr2_+_46779827
|
1.077
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr18_+_9126742
|
1.066
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr3_+_19963534
|
1.051
|
NM_004162
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr17_+_55052060
|
1.029
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr17_+_55052046
|
1.012
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr6_+_87921980
|
1.003
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr14_+_23653848
|
0.984
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_+_127629677
|
0.959
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr9_+_6747919
|
0.954
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr14_+_23654307
|
0.888
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr7_-_95789204
|
0.878
|
NM_001160210
NM_014251
|
SLC25A13
|
solute carrier family 25, member 13 (citrin)
|
|
chr7_+_140042932
|
0.875
|
NM_004546
|
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa
|
|
chrX_+_100549869
|
0.864
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr5_+_133735004
|
0.857
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr11_-_18300246
|
0.849
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chrX_+_100549885
|
0.848
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr10_+_69314347
|
0.835
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr2_+_42249896
|
0.833
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr17_+_55052065
|
0.821
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr9_-_126573349
|
0.814
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr17_+_55052185
|
0.807
|
|
|
|
|
chr12_-_44670475
|
0.793
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chrX_+_100549839
|
0.782
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_-_100549561
|
0.776
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr6_+_143423656
|
0.769
|
NM_016108
|
AIG1
|
androgen-induced 1
|
|
chrX_+_84385649
|
0.767
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr10_+_111959978
|
0.763
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr2_+_228045053
|
0.757
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr17_-_46692369
|
0.755
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr10_+_69990050
|
0.752
|
NM_030625
|
TET1
|
tet oncogene 1
|
|
chr2_+_135392857
|
0.744
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr12_+_12656022
|
0.743
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr2_+_177965734
|
0.742
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr14_+_100270222
|
0.740
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr15_+_57696016
|
0.737
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr7_-_135263737
|
0.735
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chrX_+_100549861
|
0.733
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_-_113300351
|
0.730
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr8_-_126173203
|
0.729
|
|
KIAA0196
|
KIAA0196
|
|
chr16_-_5023934
|
0.726
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr12_-_122415327
|
0.723
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr17_+_25467948
|
0.717
|
NM_032141
|
CCDC55
|
coiled-coil domain containing 55
|
|
chr2_+_219751230
|
0.717
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr3_+_19963865
|
0.715
|
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr15_+_57696200
|
0.703
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr1_-_245237925
|
0.700
|
|
ZNF695
|
zinc finger protein 695
|
|
chrX_+_30581523
|
0.696
|
|
GK
|
glycerol kinase
|
|
chr21_-_39642880
|
0.689
|
NM_004965
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr8_-_126173189
|
0.689
|
|
KIAA0196
|
KIAA0196
|
|
chr12_+_12656109
|
0.685
|
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr2_+_228045121
|
0.683
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr8_-_126173186
|
0.680
|
|
KIAA0196
|
KIAA0196
|
|
chr6_-_53321706
|
0.680
|
NM_021814
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr10_-_45410197
|
0.678
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr14_+_23653819
|
0.676
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr17_-_73636386
|
0.672
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr2_+_219751192
|
0.672
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr8_-_126173108
|
0.669
|
|
KIAA0196
|
KIAA0196
|
|
chr3_-_171382066
|
0.661
|
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr1_-_115060904
|
0.659
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr6_-_53321545
|
0.657
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr14_+_22860511
|
0.649
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr4_-_47160403
|
0.646
|
NM_017845
|
COMMD8
|
COMM domain containing 8
|
|
chr2_-_177965626
|
0.644
|
|
LOC100130691
|
hypothetical LOC100130691
|
|
chr2_-_240613480
|
0.643
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr12_+_110935618
|
0.641
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr3_-_198509276
|
0.639
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr11_-_107598472
|
0.638
|
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chrX_+_84385693
|
0.638
|
|
ZNF711
|
zinc finger protein 711
|
|
chr6_+_143423723
|
0.637
|
|
AIG1
|
androgen-induced 1
|
|
chr14_+_23653745
|
0.635
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_109810396
|
0.634
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr18_+_30875297
|
0.631
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr20_+_57948803
|
0.630
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr6_+_105511615
|
0.625
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr12_-_122415708
|
0.620
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr14_-_64040072
|
0.620
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr14_+_54807855
|
0.618
|
|
FBXO34
|
F-box protein 34
|
|
chr5_-_94916416
|
0.614
|
NM_014639
|
TTC37
|
tetratricopeptide repeat domain 37
|
|
chr5_-_167938875
|
0.613
|
|
PANK3
|
pantothenate kinase 3
|
|
chr2_+_62787534
|
0.609
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr2_+_177785667
|
0.608
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr2_-_20115269
|
0.608
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr2_+_86801081
|
0.606
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr6_-_53321658
|
0.597
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr1_+_159550789
|
0.593
|
NM_001035511
NM_001035512
NM_001035513
NM_003001
|
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa
|
|
chr11_-_118757574
|
0.592
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_77920914
|
0.592
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr13_+_48582389
|
0.590
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr4_-_113777569
|
0.589
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr4_-_186584031
|
0.589
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr11_-_82460518
|
0.588
|
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr5_-_167938991
|
0.586
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr10_+_76639917
|
0.586
|
NM_001184823
|
VDAC2
|
voltage-dependent anion channel 2
|
|
chr16_+_23754822
|
0.585
|
|
PRKCB
|
protein kinase C, beta
|
|
chr11_+_7491789
|
0.584
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr4_-_47160476
|
0.582
|
|
COMMD8
|
COMM domain containing 8
|
|
chr12_-_44670613
|
0.578
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr6_-_84994032
|
0.573
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chrX_-_7855400
|
0.568
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr11_-_82460531
|
0.566
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr14_-_35348086
|
0.566
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr10_+_70553949
|
0.565
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr14_-_19993032
|
0.564
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr3_-_151963653
|
0.562
|
NM_005067
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr3_-_151963607
|
0.561
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr7_-_103635587
|
0.561
|
NM_181747
NM_002553
|
ORC5
|
origin recognition complex, subunit 5
|
|
chr12_-_75477692
|
0.558
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr20_+_37024383
|
0.557
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr8_-_54918389
|
0.553
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr8_+_98725505
|
0.551
|
NM_178812
|
MTDH
|
metadherin
|
|
chr14_-_67353046
|
0.550
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr11_+_120828275
|
0.549
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr9_+_130749812
|
0.549
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr10_+_70553862
|
0.547
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_-_97311084
|
0.541
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr2_-_8895129
|
0.539
|
NM_020738
|
KIDINS220
|
kinase D-interacting substrate, 220kDa
|
|
chr1_-_113300138
|
0.537
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr11_-_107598538
|
0.535
|
NM_002519
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr6_+_64340388
|
0.531
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr3_-_197293377
|
0.530
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr10_+_70553936
|
0.529
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr3_-_198509808
|
0.529
|
NM_001098424
NM_004087
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr8_-_126173055
|
0.527
|
|
KIAA0196
|
KIAA0196
|
|
chr9_-_14769530
|
0.527
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr21_-_39642935
|
0.527
|
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr3_+_140545528
|
0.525
|
NM_020191
|
MRPS22
|
mitochondrial ribosomal protein S22
|
|
chr3_+_67131416
|
0.525
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr6_-_109810310
|
0.524
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr2_+_105320440
|
0.524
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr4_-_77354011
|
0.524
|
NM_005506
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr2_-_240613357
|
0.523
|
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr1_-_209818675
|
0.522
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr10_+_76256305
|
0.518
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr2_-_64734544
|
0.518
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr17_+_38247493
|
0.517
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr2_-_44076616
|
0.514
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr2_-_24869718
|
0.514
|
NM_001013663
|
C2orf79
|
chromosome 2 open reading frame 79
|
|
chr5_-_145542428
|
0.512
|
NM_020117
|
LARS
|
leucyl-tRNA synthetase
|
|
chr2_+_170259195
|
0.511
|
NM_144711
NM_001008489
|
PHOSPHO2-KLHL23
KLHL23
PHOSPHO2
|
PHOSPHO2-KLHL23 read-through transcript
kelch-like 23 (Drosophila)
phosphatase, orphan 2
|
|
chr3_-_171382213
|
0.509
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr11_-_118167073
|
0.509
|
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr2_+_86800898
|
0.507
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr11_-_118757477
|
0.506
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr5_+_65476418
|
0.505
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr1_-_33055431
|
0.505
|
|
YARS
|
tyrosyl-tRNA synthetase
|
|
chrX_+_55760774
|
0.504
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr1_-_115060857
|
0.501
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr2_-_136311219
|
0.499
|
NM_002299
|
LCT
|
lactase
|
|
chr1_+_52294554
|
0.498
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr11_-_36267550
|
0.498
|
NM_001101653
NM_014186
|
COMMD9
|
COMM domain containing 9
|
|
chr2_-_177836773
|
0.498
|
NM_001145412
NM_001145413
|
NFE2L2
|
nuclear factor (erythroid-derived 2)-like 2
|
|
chr17_+_50697355
|
0.497
|
|
HLF
|
hepatic leukemia factor
|
|
chr6_+_138230045
|
0.496
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_44076620
|
0.495
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr12_+_22669352
|
0.495
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr21_-_34909014
|
0.494
|
NM_004414
|
RCAN1
|
regulator of calcineurin 1
|
|
chr8_-_54918369
|
0.493
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_+_7491576
|
0.493
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr10_+_69314714
|
0.492
|
|
SIRT1
|
sirtuin 1
|
|
chr1_-_92724181
|
0.492
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr2_+_86800940
|
0.491
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr7_-_87343599
|
0.490
|
NM_018843
|
SLC25A40
|
solute carrier family 25, member 40
|
|
chr14_-_22468445
|
0.490
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr2_-_176575171
|
0.487
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chrX_+_146801236
|
0.487
|
|
FMR1
|
fragile X mental retardation 1
|
|
chr1_+_200191241
|
0.486
|
NM_006335
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast)
|
|
chr7_+_96473225
|
0.484
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr14_-_34661156
|
0.484
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr4_+_113777568
|
0.482
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr5_-_137396677
|
0.482
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr17_+_44429748
|
0.482
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr14_-_54438913
|
0.482
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr2_-_21081694
|
0.481
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr2_-_25328452
|
0.481
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chrX_-_76928142
|
0.480
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr6_-_122834580
|
0.479
|
NM_020755
|
SERINC1
|
serine incorporator 1
|
|
chr2_-_171725604
|
0.478
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr8_-_126173227
|
0.477
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr8_+_67787444
|
0.475
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr6_+_155579788
|
0.474
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr18_+_30810889
|
0.473
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr7_-_105712565
|
0.473
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr6_-_112515329
|
0.473
|
NM_016262
|
TUBE1
|
tubulin, epsilon 1
|
|
chr10_-_3817205
|
0.472
|
|
KLF6
|
Kruppel-like factor 6
|
|
chrX_+_47305459
|
0.472
|
NM_001654
|
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog
|
|
chr13_-_49597776
|
0.471
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|