|
chr2_-_189752575
|
0.969
|
NM_000393
|
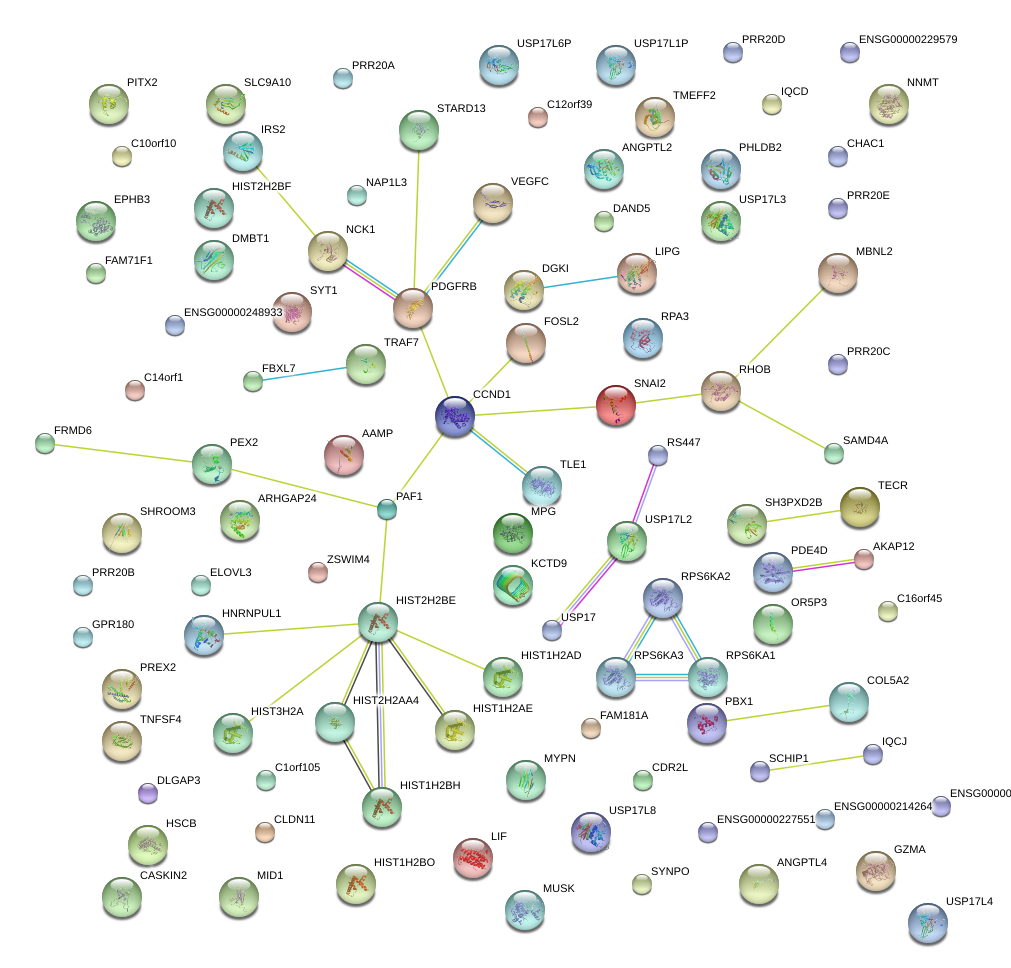
COL5A2
|
collagen, type V, alpha 2
|
|
chr17_+_70495318
|
0.966
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr6_-_166960690
|
0.805
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr22_-_28972683
|
0.786
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr2_+_20510312
|
0.758
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr10_+_124310170
|
0.634
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr3_+_112934016
|
0.630
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr13_-_109236897
|
0.556
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr12_+_77782719
|
0.525
|
|
SYT1
|
synaptotagmin I
|
|
chr5_-_171814026
|
0.523
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr6_-_166960649
|
0.516
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr5_-_149515613
|
0.495
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_+_86968069
|
0.459
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_+_54434230
|
0.444
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr5_+_15553288
|
0.432
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chrX_-_10605726
|
0.423
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr14_+_54104383
|
0.421
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chrX_-_92815222
|
0.419
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr9_+_112470871
|
0.408
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr3_+_161040343
|
0.406
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr10_+_69539255
|
0.397
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr9_-_83494198
|
0.385
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr14_+_51025604
|
0.372
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr3_+_185762233
|
0.369
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr6_-_166960496
|
0.367
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr18_+_45342398
|
0.363
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_+_162795328
|
0.363
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_-_35143570
|
0.360
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr6_+_151603264
|
0.355
|
|
|
|
|
chr11_+_113671744
|
0.348
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr6_+_151603201
|
0.340
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr9_-_128910132
|
0.337
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr5_-_171813960
|
0.332
|
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr8_+_69027143
|
0.331
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr2_-_192767844
|
0.330
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr5_+_150008030
|
0.328
|
|
SYNPO
|
synaptopodin
|
|
chr14_+_93455010
|
0.325
|
NM_138344
|
FAM181A
|
family with sequence similarity 181, member A
|
|
chr14_-_75196911
|
0.325
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr2_-_218843109
|
0.314
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr19_+_13767207
|
0.312
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr1_-_171442937
|
0.311
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr4_-_177950658
|
0.304
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr2_-_218843088
|
0.298
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr15_+_39032927
|
0.296
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr13_-_32757835
|
0.294
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr10_+_103976132
|
0.293
|
NM_152310
|
ELOVL3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3
|
|
chr16_+_2145810
|
0.292
|
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr1_+_170656450
|
0.289
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr5_-_149515502
|
0.286
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr3_+_161040439
|
0.286
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_-_7804094
|
0.278
|
NM_153445
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3
|
|
chr16_+_2145765
|
0.277
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr10_-_44794146
|
0.269
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr19_+_12941431
|
0.263
|
NM_152654
|
DAND5
|
DAN domain family, member 5
|
|
chr4_+_8935987
|
0.260
|
NM_001105662
|
USP17L2
USP17L6P
USP17
|
ubiquitin specific peptidase 17-like 2
ubiquitin specific peptidase 17-like 6 (pseudogene)
ubiquitin specific peptidase 17
|
|
chr1_-_226712140
|
0.251
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr8_+_69026895
|
0.250
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr22_+_27468018
|
0.248
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr13_+_56619622
|
0.248
|
NM_198441
NM_001130404
NM_001130405
NM_001130406
NM_001130407
|
PRR20A
PRR20B
PRR20C
PRR20D
PRR20E
|
proline rich 20A
proline rich 20B
proline rich 20C
proline rich 20D
proline rich 20E
|
|
chr7_-_77163475
|
0.243
|
|
|
|
|
chr6_+_151603346
|
0.239
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_-_44573343
|
0.238
|
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr3_-_113495763
|
0.238
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr5_-_149515331
|
0.237
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_-_111777956
|
0.237
|
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_+_128136347
|
0.237
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr17_-_71023221
|
0.236
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr2_-_192767494
|
0.233
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr7_-_7646573
|
0.230
|
|
RPA3
|
replication protein A3, 14kDa
|
|
chr1_-_147665783
|
0.225
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr13_+_94052022
|
0.224
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr10_-_102880892
|
0.218
|
NM_001085398
|
TLX1NB
|
TLX1 neighbor
|
|
chr19_+_46462113
|
0.218
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr11_+_69176391
|
0.218
|
|
CCND1
|
cyclin D1
|
|
chr2_+_28469850
|
0.216
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr8_-_49996353
|
0.215
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr12_-_112143209
|
0.215
|
NM_138451
|
IQCD
|
IQ motif containing D
|
|
chr19_+_14501406
|
0.214
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr8_-_25371858
|
0.212
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr17_-_1566223
|
0.209
|
|
C17orf91
|
chromosome 17 open reading frame 91
|
|
chr12_+_21570457
|
0.209
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr5_-_58607673
|
0.208
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_+_15503623
|
0.205
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr4_+_77575276
|
0.199
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr7_-_137182125
|
0.198
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr19_+_14501403
|
0.197
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr3_+_171621721
|
0.196
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr3_+_138132006
|
0.195
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr13_+_96726415
|
0.194
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr12_+_77782648
|
0.189
|
|
SYT1
|
synaptotagmin I
|
|
chr11_-_85074848
|
0.189
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr15_+_41597097
|
0.189
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr19_+_46461897
|
0.184
|
NM_007040
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr10_+_102881050
|
0.184
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chr14_-_103251497
|
0.182
|
NM_001100118
NM_001100119
NM_005432
|
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3
|
|
chr19_-_14501048
|
0.181
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr5_-_16795336
|
0.175
|
|
MYO10
|
myosin X
|
|
chr7_-_121571472
|
0.174
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr11_-_27679175
|
0.174
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_201729893
|
0.174
|
NM_014359
|
OPTC
|
opticin
|
|
chr17_-_43977273
|
0.169
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr12_-_54409710
|
0.169
|
|
CD63
|
CD63 molecule
|
|
chr17_-_71023048
|
0.168
|
|
CASKIN2
|
CASK interacting protein 2
|
|
chr4_-_140696740
|
0.168
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr19_+_14501363
|
0.161
|
NM_138501
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr9_+_36180927
|
0.161
|
|
CLTA
|
clathrin, light chain A
|
|
chr17_-_36436979
|
0.161
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr2_+_216695062
|
0.160
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chr19_-_44573468
|
0.160
|
NM_019088
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr17_-_36456949
|
0.159
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr11_+_113436438
|
0.159
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr17_+_54763689
|
0.158
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr15_-_84139021
|
0.158
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr16_+_1772929
|
0.158
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr19_-_59355246
|
0.157
|
NM_024316
|
LENG1
|
leukocyte receptor cluster (LRC) member 1
|
|
chr9_-_72926333
|
0.157
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr1_-_120285753
|
0.154
|
|
NOTCH2
|
notch 2
|
|
chr4_+_176076083
|
0.152
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr3_+_138063697
|
0.151
|
NM_006153
|
NCK1
|
NCK adaptor protein 1
|
|
chr10_+_17310263
|
0.151
|
NM_003380
|
VIM
|
vimentin
|
|
chr11_-_102174102
|
0.151
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chrX_+_46581418
|
0.148
|
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr5_+_15553539
|
0.141
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr3_+_131175803
|
0.140
|
NM_007117
|
TRH
|
thyrotropin-releasing hormone
|
|
chr2_+_71534260
|
0.139
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr16_+_1773004
|
0.136
|
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr1_+_16220952
|
0.135
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr7_-_526005
|
0.135
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr12_+_47945132
|
0.135
|
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr9_+_36180936
|
0.134
|
|
CLTA
|
clathrin, light chain A
|
|
chr14_+_64041257
|
0.132
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr7_-_27108826
|
0.131
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr22_+_34025872
|
0.130
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr10_-_79067296
|
0.130
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chrX_-_145703940
|
0.130
|
NM_001144064
|
CXorf51
|
chromosome X open reading frame 51
|
|
chr10_+_99390459
|
0.129
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr4_+_154083584
|
0.129
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr16_+_67778441
|
0.126
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr2_+_12774264
|
0.126
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr9_+_36180885
|
0.126
|
|
CLTA
|
clathrin, light chain A
|
|
chr16_+_81218142
|
0.125
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr10_+_114699953
|
0.124
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr8_+_69027166
|
0.124
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr11_-_102174032
|
0.124
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr3_-_50340598
|
0.124
|
|
TUSC2
|
tumor suppressor candidate 2
|
|
chr16_+_3433606
|
0.124
|
NM_001083601
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr4_-_70115037
|
0.123
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr14_-_91642703
|
0.123
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr11_-_129803515
|
0.122
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr12_+_27568311
|
0.121
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr12_+_47009029
|
0.120
|
NM_181788
|
H1FNT
|
H1 histone family, member N, testis-specific
|
|
chr9_-_98421894
|
0.120
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chrX_-_10548458
|
0.120
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_-_7471832
|
0.120
|
NM_133491
|
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2
|
|
chr3_-_71196696
|
0.119
|
|
FOXP1
|
forkhead box P1
|
|
chr9_-_14303509
|
0.119
|
|
NFIB
|
nuclear factor I/B
|
|
chr9_+_36180956
|
0.117
|
|
CLTA
|
clathrin, light chain A
|
|
chr19_-_54646837
|
0.116
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr21_-_46473143
|
0.116
|
NM_001001438
NM_001145436
NM_001145437
NM_002340
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr21_-_35174742
|
0.116
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_+_97856148
|
0.116
|
|
|
|
|
chr9_-_106407728
|
0.115
|
NM_001004481
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2
|
|
chr3_-_161766055
|
0.115
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr19_-_63301386
|
0.115
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr3_-_150993299
|
0.114
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr6_+_26212082
|
0.113
|
NM_003542
|
HIST1H4C
|
histone cluster 1, H4c
|
|
chr1_+_216585298
|
0.112
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr3_-_161766005
|
0.112
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr19_-_3928840
|
0.111
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr17_-_77411803
|
0.111
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr13_+_96726197
|
0.110
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_+_90711887
|
0.110
|
|
LOC100129716
|
hypothetical LOC100129716
|
|
chr2_-_162808123
|
0.109
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr7_-_141320041
|
0.109
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr3_+_130641779
|
0.108
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr9_+_129604962
|
0.108
|
NM_004957
|
FPGS
|
folylpolyglutamate synthase
|
|
chr13_+_26029799
|
0.107
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr19_+_46462169
|
0.107
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr3_+_162697289
|
0.106
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr11_+_19328846
|
0.105
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr3_+_52786642
|
0.105
|
NM_002215
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr18_-_45272840
|
0.104
|
NM_000985
NM_001035006
|
RPL17
|
ribosomal protein L17
|
|
chr8_-_23768242
|
0.104
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_-_158704056
|
0.103
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr13_-_95094920
|
0.103
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr4_-_118226119
|
0.103
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr3_-_21767819
|
0.103
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr4_+_81325447
|
0.103
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr6_-_63053811
|
0.102
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr16_+_81218074
|
0.102
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr18_-_45272820
|
0.102
|
|
RPL17
|
ribosomal protein L17
|
|
chr3_+_130641828
|
0.101
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr22_-_41741100
|
0.100
|
NM_001184970
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr7_-_143230096
|
0.100
|
NM_014719
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr22_+_34025792
|
0.099
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr18_+_58005519
|
0.098
|
|
KIAA1468
|
KIAA1468
|
|
chr14_-_23653965
|
0.098
|
|
NRL
|
neural retina leucine zipper
|
|
chr12_+_47945086
|
0.098
|
NM_032704
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr12_-_90097384
|
0.097
|
NM_133503
|
DCN
|
decorin
|
|
chr4_-_83771386
|
0.097
|
|
SCD5
|
stearoyl-CoA desaturase 5
|