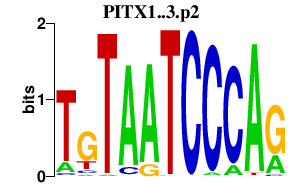
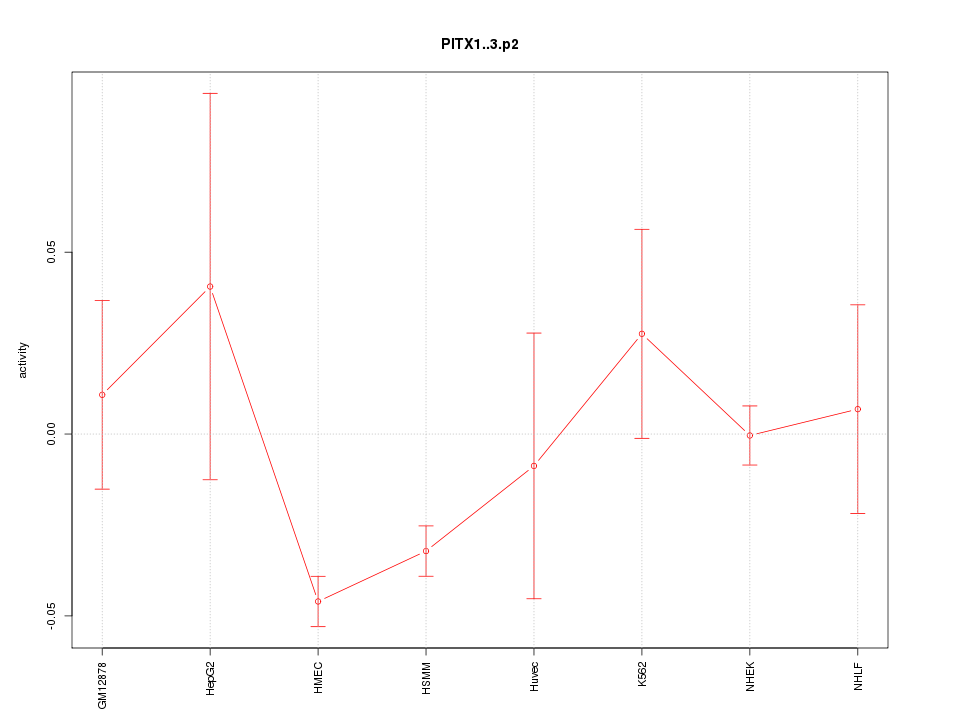
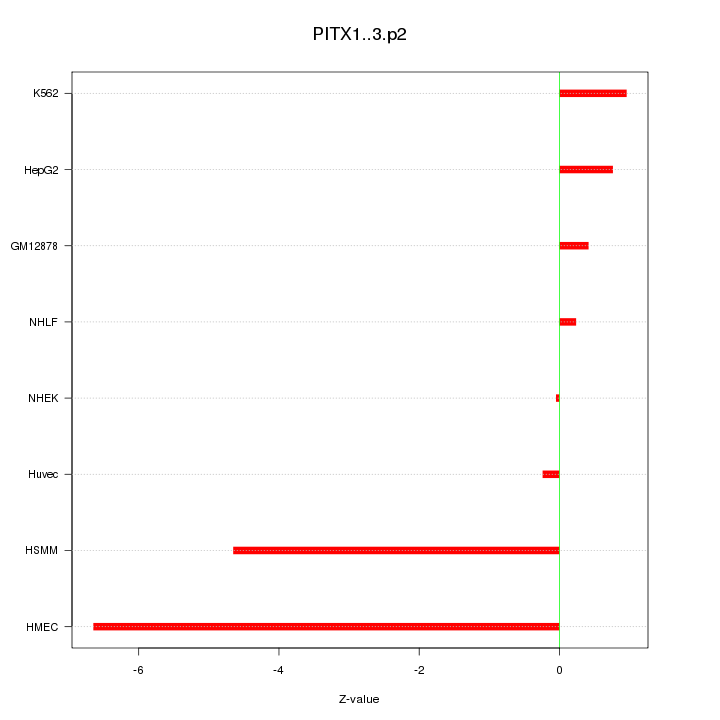
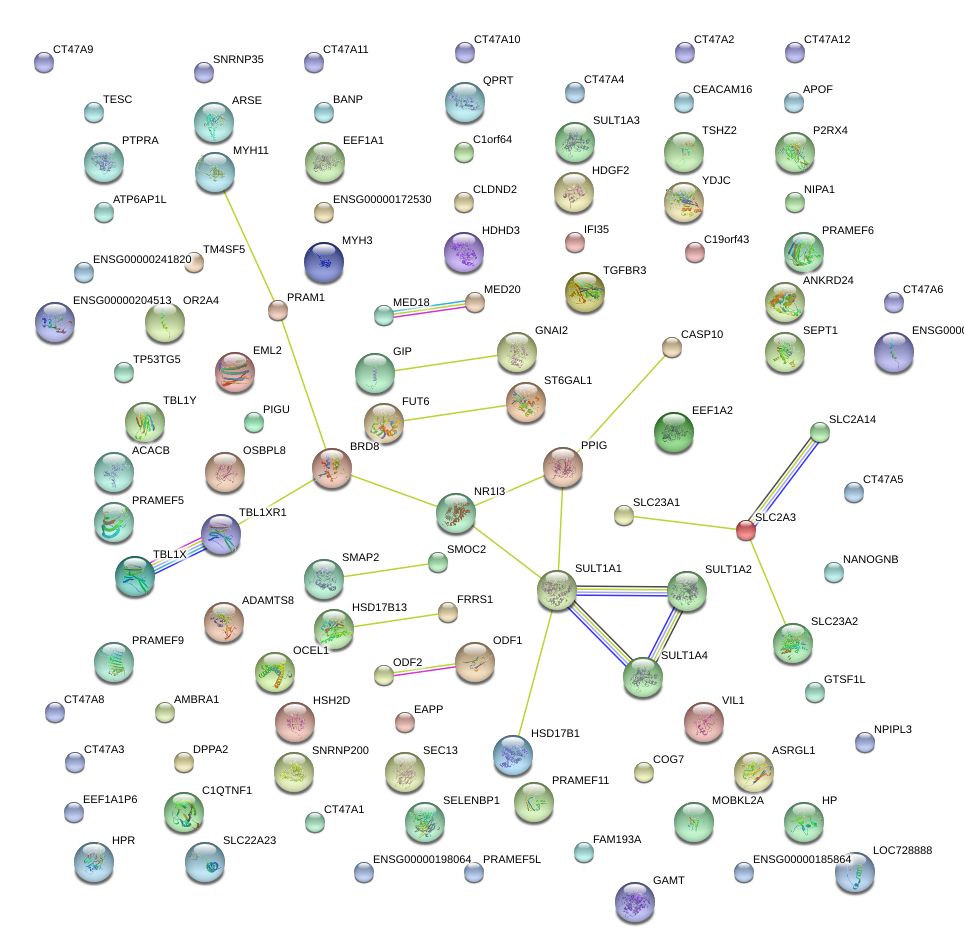
|
chr1_-_154536051
|
5.603
|
NM_001004319
|
VHLL
|
von Hippel-Lindau tumor suppressor-like
|
|
chr17_-_59818033
|
3.877
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr1_-_92144146
|
3.609
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr20_+_60946920
|
3.483
|
NM_080750
|
DPH3P1
|
DPH3, KTI11 homolog (S. cerevisiae) pseudogene 1
|
|
chrX_-_2892288
|
3.228
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr1_-_100003936
|
2.790
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr12_+_122510338
|
2.632
|
NM_180699
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12)
|
|
chr16_-_21762256
|
2.614
|
|
LOC100132247
NPIPL3
LOC613037
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
nuclear pore complex interacting protein pseudogene
|
|
chr1_-_149611768
|
2.499
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr12_-_116021473
|
2.481
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr1_+_16203317
|
2.446
|
NM_178840
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr16_+_70654625
|
2.352
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr16_+_29378694
|
2.294
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr16_+_30118036
|
2.281
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr19_-_56564068
|
2.260
|
NM_152353
|
CLDND2
|
claudin domain containing 2
|
|
chr17_+_4621924
|
2.181
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr7_-_143587653
|
2.107
|
NM_001005328
|
OR2A7
OR2A4
|
olfactory receptor, family 2, subfamily A, member 7
olfactory receptor, family 2, subfamily A, member 4
|
|
chr16_+_29597829
|
2.106
|
NM_014298
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr11_+_61861458
|
2.105
|
|
ASRGL1
|
asparaginase like 1
|
|
chr19_+_16115454
|
2.097
|
|
HSH2D
|
hematopoietic SH2 domain containing
|
|
chr12_+_108149558
|
2.097
|
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr16_+_86561124
|
2.026
|
NM_001173543
|
BANP
|
BTG3 associated nuclear protein
|
|
chr3_-_110517929
|
2.007
|
NM_138815
|
DPPA2
|
developmental pluripotency associated 2
|
|
chrX_+_9391314
|
1.978
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_50834483
|
1.952
|
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr19_+_4134350
|
1.942
|
NM_133475
|
ANKRD24
|
ankyrin repeat domain 24
|
|
chr17_-_44400952
|
1.940
|
NM_004123
|
GIP
|
gastric inhibitory polypeptide
|
|
chr22_-_20314274
|
1.911
|
NM_001017964
|
YDJC
|
YdjC homolog (bacterial)
|
|
chr19_+_17198012
|
1.889
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr4_+_2596956
|
1.884
|
NM_003704
|
FAM193A
|
family with sequence similarity 193, member A
|
|
chr9_-_115178161
|
1.875
|
NM_031219
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr1_-_159474503
|
1.869
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr20_+_2851852
|
1.862
|
NM_080841
|
PTPRA
|
protein tyrosine phosphatase, receptor type, A
|
|
chr16_-_15741438
|
1.858
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr22_-_20314366
|
1.851
|
|
YDJC
|
YdjC homolog (bacterial)
|
|
chr12_-_55042793
|
1.814
|
NM_001638
|
APOF
|
apolipoprotein F
|
|
chr17_+_37957509
|
1.775
|
NM_000413
|
HSD17B1
|
hydroxysteroid (17-beta) dehydrogenase 1
|
|
chr6_-_3390197
|
1.773
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr11_-_46569489
|
1.758
|
NM_017749
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr16_-_18705578
|
1.727
|
|
|
|
|
chr19_-_5790702
|
1.704
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr16_-_23371952
|
1.695
|
NM_153603
|
COG7
|
component of oligomeric golgi complex 7
|
|
chr9_+_134884627
|
1.695
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_+_40631603
|
1.693
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr17_-_10500189
|
1.670
|
NM_002470
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic
|
|
chr19_+_4439690
|
1.667
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr9_+_130257254
|
1.666
|
NM_153436
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr14_-_34078532
|
1.665
|
NM_018453
|
EAPP
|
E2F-associated phosphoprotein
|
|
chr20_-_43440290
|
1.644
|
NM_014477
|
TP53TG5
|
TP53 target 5
|
|
chr19_-_8473473
|
1.641
|
NM_032152
|
PRAM1
|
PML-RARA regulated adaptor molecule 1
|
|
chr2_+_201755865
|
1.637
|
NM_001230
NM_032974
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr17_+_38412267
|
1.625
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr3_+_188222337
|
1.620
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr2_+_218992053
|
1.609
|
NM_007127
|
VIL1
|
villin 1
|
|
chr1_+_13232405
|
1.600
|
NM_001013407
NM_001010889
|
PRAMEF5
PRAMEF6
|
PRAME family member 5
PRAME family member 6
|
|
chr19_-_1352474
|
1.587
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr20_-_32728749
|
1.585
|
NM_080476
|
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U
|
|
chr12_-_7935010
|
1.583
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr12_+_120132233
|
1.557
|
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr9_+_131291144
|
1.553
|
|
|
|
|
chr3_-_10337837
|
1.549
|
NM_183352
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr20_-_41788975
|
1.518
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr16_-_30301634
|
1.512
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr3_-_10337724
|
1.512
|
NM_001136232
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr6_-_132064233
|
1.510
|
NM_030908
|
OR2A4
|
olfactory receptor, family 2, subfamily A, member 4
|
|
chr1_+_28528099
|
1.508
|
NM_001127350
NM_017638
|
MED18
|
mediator complex subunit 18
|
|
chr15_-_70838703
|
1.503
|
|
|
|
|
chr2_-_96314148
|
1.491
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr20_-_4938913
|
1.489
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr19_-_2036227
|
1.489
|
|
MOBKL2A
|
MOB1, Mps One Binder kinase activator-like 2A (yeast)
|
|
chr19_-_12706458
|
1.478
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr19_+_49894260
|
1.475
|
NM_001039213
|
CEACAM16
|
carcinoembryonic antigen-related cell adhesion molecule 16
|
|
chr4_-_88463031
|
1.461
|
NM_001136230
NM_178135
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr5_+_81636921
|
1.456
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr15_-_20638283
|
1.454
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr20_+_51235228
|
1.452
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr12_+_7809078
|
1.440
|
NM_001145465
|
NANOGNB
|
NANOG neighbor homeobox
|
|
chrX_-_119898692
|
1.438
|
NM_173571
NM_001080137
NM_001080138
NM_001080139
NM_001080141
NM_001080142
NM_001080143
NM_001080144
NM_001080145
NM_001080146
|
CT47A11
CT47A10
CT47A9
CT47A8
CT47A6
CT47A5
CT47A4
CT47A3
CT47A2
CT47A1
|
cancer/testis antigen family 47, member A11
cancer/testis antigen family 47, member A10
cancer/testis antigen family 47, member A9
cancer/testis antigen family 47, member A8
cancer/testis antigen family 47, member A6
cancer/testis antigen family 47, member A5
cancer/testis antigen family 47, member A4
cancer/testis antigen family 47, member A3
cancer/testis antigen family 47, member A2
cancer/testis antigen family 47, member A1
|
|
chr19_-_12706480
|
1.420
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr7_+_821719
|
1.416
|
NM_001171946
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr17_+_1612031
|
1.389
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr17_+_46176105
|
1.367
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr17_-_78599695
|
1.367
|
|
B3GNTL1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1
|
|
chr6_+_42126242
|
1.360
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr2_+_38746555
|
1.359
|
NM_138801
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr22_-_29231597
|
1.358
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr19_+_6323443
|
1.344
|
NM_032306
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr17_+_1612056
|
1.341
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr17_+_70997280
|
1.335
|
|
KIAA0195
|
KIAA0195
|
|
chr12_+_31691360
|
1.329
|
NM_001135864
|
METTL20
|
methyltransferase like 20
|
|
chr19_-_54857003
|
1.323
|
|
IRF3
|
interferon regulatory factor 3
|
|
chr7_+_73262188
|
1.322
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr19_-_44497815
|
1.319
|
NM_020862
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1
|
|
chr15_-_20666684
|
1.317
|
|
LOC283683
|
hypothetical LOC283683
|
|
chr16_-_86527608
|
1.314
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr1_-_149245503
|
1.310
|
NM_001163259
NM_001163260
NM_001040217
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr4_+_74499651
|
1.303
|
|
ALB
|
albumin
|
|
chr19_-_59383944
|
1.297
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr9_+_131605270
|
1.294
|
|
TOR1B
|
torsin family 1, member B (torsin B)
|
|
chr19_+_52532210
|
1.291
|
NM_018485
|
GPR77
|
G protein-coupled receptor 77
|
|
chr22_-_22714230
|
1.285
|
NM_000853
|
GSTT1
|
glutathione S-transferase theta 1
|
|
chr6_-_150001376
|
1.284
|
NM_007044
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1
|
|
chr9_+_131605215
|
1.280
|
NM_014506
|
TOR1B
|
torsin family 1, member B (torsin B)
|
|
chr12_-_131882616
|
1.264
|
|
|
|
|
chr5_-_10361167
|
1.261
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr20_-_32354736
|
1.260
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr8_-_141537859
|
1.259
|
NM_031466
|
TRAPPC9
|
trafficking protein particle complex 9
|
|
chr7_-_100645032
|
1.246
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr19_+_12064077
|
1.239
|
|
ZNF788
|
zinc finger family member 788
|
|
chrX_-_70243362
|
1.236
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr15_-_39623750
|
1.234
|
NM_015540
|
RPAP1
|
RNA polymerase II associated protein 1
|
|
chr20_-_47307554
|
1.231
|
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr22_+_20350272
|
1.223
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr16_-_88623809
|
1.222
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr17_+_73694992
|
1.222
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr20_+_43952189
|
1.218
|
|
CTSA
|
cathepsin A
|
|
chr11_-_64520964
|
1.215
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr19_+_51059357
|
1.215
|
NM_004497
|
FOXA3
|
forkhead box A3
|
|
chr20_+_35240936
|
1.213
|
|
RPN2
|
ribophorin II
|
|
chr22_+_19248733
|
1.211
|
|
MED15
|
mediator complex subunit 15
|
|
chr16_+_70599987
|
1.201
|
NM_001361
|
DHODH
|
dihydroorotate dehydrogenase
|
|
chr22_-_36570287
|
1.199
|
NM_138797
|
ANKRD54
|
ankyrin repeat domain 54
|
|
chr16_-_2254318
|
1.196
|
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr9_-_116196505
|
1.193
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr17_+_29707583
|
1.182
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr1_+_10432362
|
1.181
|
NM_001302
|
CORT
|
cortistatin
|
|
chr4_+_56731117
|
1.158
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chr7_-_2561853
|
1.152
|
NM_152743
|
BAAT1
|
BRCA1-associated ATM activator 1
|
|
chr6_-_31871530
|
1.151
|
NM_006295
|
VARS
|
valyl-tRNA synthetase
|
|
chr16_-_24934151
|
1.140
|
NM_001006634
NM_018054
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr17_+_1611983
|
1.138
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr3_+_52296893
|
1.138
|
|
GLYCTK
|
glycerate kinase
|
|
chr18_-_3001944
|
1.132
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr19_-_56825400
|
1.128
|
NM_003830
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5
|
|
chr16_-_31013626
|
1.122
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr16_-_27187559
|
1.120
|
NM_145080
|
NSMCE1
|
non-SMC element 1 homolog (S. cerevisiae)
|
|
chr9_+_130504591
|
1.119
|
NM_013355
|
PKN3
|
protein kinase N3
|
|
chr7_+_44007012
|
1.111
|
NM_175064
|
SPDYE1
|
speedy homolog E1 (Xenopus laevis)
|
|
chr19_+_60168463
|
1.104
|
NM_001174081
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr2_+_201756048
|
1.083
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr6_+_31706345
|
1.083
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr11_+_22652894
|
1.074
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr16_-_73798539
|
1.072
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr9_+_6710862
|
1.070
|
NM_001146696
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr19_+_53656317
|
1.069
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr20_+_309272
|
1.067
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr22_+_40372957
|
1.054
|
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr17_+_59244363
|
1.047
|
|
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42
|
|
chr1_+_24713396
|
1.046
|
|
RCAN3
|
RCAN family member 3
|
|
chr22_+_17012841
|
1.039
|
|
USP18
|
ubiquitin specific peptidase 18
|
|
chr1_-_17253073
|
1.033
|
NM_003000
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr12_+_120132319
|
1.033
|
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr3_+_4510160
|
1.028
|
|
ITPR1
|
inositol 1,4,5-triphosphate receptor, type 1
|
|
chr17_-_38427957
|
1.026
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr19_+_50100843
|
1.021
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr7_+_65217239
|
1.017
|
NM_001040647
NM_001040648
NM_001142414
NM_014478
|
CRCP
|
CGRP receptor component
|
|
chr7_-_129478430
|
1.006
|
NM_016478
|
ZC3HC1
|
zinc finger, C3HC-type containing 1
|
|
chr19_-_16111695
|
1.006
|
|
|
|
|
chr16_+_65558878
|
1.006
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr12_-_52975777
|
1.004
|
NM_006163
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr17_+_68740952
|
1.002
|
|
C17orf80
|
chromosome 17 open reading frame 80
|
|
chr20_+_35351473
|
1.001
|
|
MANBAL
|
mannosidase, beta A, lysosomal-like
|
|
chr19_-_59542197
|
0.997
|
NM_012276
|
LILRA4
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4
|
|
chr1_+_28029909
|
0.988
|
|
PPP1R8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr12_-_52975823
|
0.988
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr19_-_38052481
|
0.988
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr22_+_34109095
|
0.983
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr17_+_17320989
|
0.982
|
NM_018019
|
MED9
|
mediator complex subunit 9
|
|
chr11_+_22652957
|
0.981
|
|
GAS2
|
growth arrest-specific 2
|
|
chrX_-_53646332
|
0.980
|
|
HUWE1
|
HECT, UBA and WWE domain containing 1
|
|
chr19_+_50234135
|
0.979
|
NM_007056
|
CLASRP
|
CLK4-associating serine/arginine rich protein
|
|
chr20_+_32598352
|
0.976
|
NM_181509
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr20_+_35351428
|
0.976
|
NM_001003897
NM_022077
|
MANBAL
|
mannosidase, beta A, lysosomal-like
|
|
chr20_+_43423990
|
0.971
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr17_-_59451662
|
0.970
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr11_+_69872033
|
0.963
|
|
PPFIA1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1
|
|
chr19_+_17906904
|
0.962
|
NM_138442
|
CCDC124
|
coiled-coil domain containing 124
|
|
chr6_-_35588613
|
0.962
|
NM_003322
|
TULP1
|
tubby like protein 1
|
|
chr1_-_152847105
|
0.953
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr16_+_29597965
|
0.948
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr1_+_28029922
|
0.948
|
|
PPP1R8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr5_-_134291882
|
0.945
|
|
|
|
|
chr2_+_38746742
|
0.945
|
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr1_-_153379344
|
0.945
|
NM_018973
NM_153741
|
DPM3
|
dolichyl-phosphate mannosyltransferase polypeptide 3
|
|
chr16_+_43009
|
0.943
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr4_-_23500706
|
0.942
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_+_42126228
|
0.939
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr16_+_65552600
|
0.936
|
NM_001185177
NM_024922
|
CES3
|
carboxylesterase 3
|
|
chr1_+_154605626
|
0.935
|
NM_020407
|
RHBG
|
Rh family, B glycoprotein (gene/pseudogene)
|
|
chr11_-_65244718
|
0.934
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr19_-_12858956
|
0.922
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr16_-_73576508
|
0.920
|
NM_030581
|
WDR59
|
WD repeat domain 59
|
|
chr16_-_57309625
|
0.918
|
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr19_-_44518465
|
0.918
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr9_+_131428225
|
0.917
|
NM_014064
|
METTL11A
|
methyltransferase like 11A
|
|
chr19_-_60528480
|
0.912
|
NM_001085488
|
TMEM150B
|
transmembrane protein 150B
|
|
chr19_+_55383249
|
0.912
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr1_-_152847256
|
0.909
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr22_+_19458378
|
0.908
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chrX_+_96025562
|
0.906
|
NM_013347
|
RPA4
|
replication protein A4, 30kDa
|