|
chr16_+_23754772
|
6.703
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr16_+_23754822
|
5.439
|
|
PRKCB
|
protein kinase C, beta
|
|
chr16_+_23754860
|
5.290
|
|
PRKCB
|
protein kinase C, beta
|
|
chr14_-_106084519
|
5.066
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_23754997
|
4.439
|
|
PRKCB
|
protein kinase C, beta
|
|
chr6_+_26291936
|
4.146
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr1_+_226712430
|
3.561
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr1_-_153098811
|
3.021
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_+_50318801
|
2.963
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr3_+_99733432
|
2.885
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr6_-_33016561
|
2.811
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr6_-_27968906
|
2.728
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr22_+_21065134
|
2.638
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr6_-_26151863
|
2.313
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr17_-_16816113
|
2.288
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr1_+_205736095
|
2.264
|
NM_000573
NM_000651
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr10_+_124897627
|
1.957
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr6_-_26232109
|
1.913
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr22_+_20316968
|
1.910
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr15_+_81906983
|
1.818
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr6_+_26266311
|
1.791
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr17_-_59363352
|
1.755
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr12_+_56290229
|
1.737
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr8_+_19840861
|
1.685
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr6_+_27222839
|
1.621
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr5_-_66528330
|
1.582
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr6_+_27969181
|
1.564
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr12_+_91620749
|
1.498
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr5_-_131907071
|
1.465
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr9_-_126573349
|
1.436
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr3_+_114733907
|
1.436
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr22_+_21370394
|
1.397
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr12_-_10953427
|
1.381
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr22_+_21042091
|
1.370
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr6_+_10693964
|
1.332
|
NM_145655
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr5_-_88155360
|
1.325
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr22_+_21094094
|
1.300
|
|
|
|
|
chr8_+_56954899
|
1.292
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr6_+_26232351
|
1.290
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr19_+_14554895
|
1.263
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr15_+_81907176
|
1.258
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr6_-_26324850
|
1.249
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr16_-_87280380
|
1.248
|
NM_178310
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr19_-_18253413
|
1.234
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr6_+_32515596
|
1.218
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr8_+_56954946
|
1.215
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr10_-_64246129
|
1.214
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr21_-_31123797
|
1.211
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr22_+_20880159
|
1.203
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr13_-_45640679
|
1.189
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_-_27222556
|
1.182
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr1_-_148124738
|
1.179
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr10_-_64246080
|
1.167
|
|
EGR2
|
early growth response 2
|
|
chr6_+_27323458
|
1.158
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr9_-_139316523
|
1.139
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr8_+_19841010
|
1.127
|
|
LPL
|
lipoprotein lipase
|
|
chr22_+_20715340
|
1.125
|
|
|
|
|
chr6_-_27208507
|
1.123
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr6_+_27208763
|
1.122
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr12_+_111829121
|
1.118
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr6_+_25834983
|
1.074
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr6_-_27890496
|
1.068
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr6_-_168219380
|
1.065
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr1_+_202000906
|
1.064
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr1_+_148089251
|
1.059
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr19_+_3523958
|
1.056
|
|
HMG20B
|
high-mobility group 20B
|
|
chr17_+_19999784
|
1.050
|
NM_001033554
NM_001033555
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr17_-_44249379
|
1.049
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr19_-_18253048
|
1.041
|
|
JUND
|
jun D proto-oncogene
|
|
chr6_+_135558572
|
1.038
|
|
|
|
|
chr15_+_76171981
|
1.032
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr10_-_98021144
|
1.031
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr6_+_27941012
|
1.031
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr19_+_60803412
|
1.031
|
NM_153219
|
ZNF524
|
zinc finger protein 524
|
|
chr14_-_105763152
|
1.028
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr22_+_21495273
|
1.020
|
|
|
|
|
chr1_+_5975286
|
1.018
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr22_-_22426551
|
1.013
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr1_+_5975345
|
1.009
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr7_+_142629251
|
1.002
|
NM_176882
|
TAS2R40
|
taste receptor, type 2, member 40
|
|
chr19_-_51856129
|
0.982
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr6_+_37245860
|
0.978
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_-_18369407
|
0.976
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr5_-_176868483
|
0.953
|
|
DOK3
|
docking protein 3
|
|
chr6_+_26359805
|
0.950
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr6_+_27883877
|
0.943
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr2_-_2313894
|
0.935
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr6_+_34312979
|
0.932
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_+_20880112
|
0.931
|
|
|
|
|
chr22_+_44059491
|
0.927
|
NM_001167574
NM_006953
|
UPK3A
|
uroplakin 3A
|
|
chr2_+_96790298
|
0.927
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr11_-_31789431
|
0.926
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr22_+_31080871
|
0.925
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr15_-_38953628
|
0.923
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr14_-_106184988
|
0.920
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_-_80344096
|
0.903
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr1_-_148126089
|
0.898
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr7_+_16759845
|
0.880
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chrX_+_12903145
|
0.878
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr6_-_168222645
|
0.872
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr21_-_38415273
|
0.855
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr22_+_21407066
|
0.850
|
|
|
|
|
chr6_+_34312906
|
0.847
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_17377502
|
0.842
|
|
LOC100507535
|
hypothetical LOC100507535
|
|
chr4_-_40212670
|
0.840
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr2_-_157890400
|
0.840
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr7_+_100021862
|
0.831
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr11_+_27019084
|
0.828
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr16_-_87280289
|
0.827
|
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr3_+_153074118
|
0.823
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr2_-_219614481
|
0.817
|
NM_152389
NM_194302
|
CCDC108
|
coiled-coil domain containing 108
|
|
chr17_+_60563963
|
0.811
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr6_+_26381122
|
0.804
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr14_-_105862080
|
0.802
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr3_+_188130967
|
0.795
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr6_+_97117144
|
0.795
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr8_+_56954953
|
0.782
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr11_-_558419
|
0.770
|
|
|
|
|
chr19_-_18253259
|
0.762
|
|
|
|
|
chr19_+_59820413
|
0.758
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr8_+_19841209
|
0.755
|
|
LPL
|
lipoprotein lipase
|
|
chr19_+_3523950
|
0.754
|
|
HMG20B
|
high-mobility group 20B
|
|
chr6_+_34312667
|
0.752
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_26141774
|
0.742
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr6_-_89984214
|
0.741
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr4_-_71751070
|
0.729
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr12_+_111829191
|
0.704
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chrX_+_37524250
|
0.704
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr19_-_19142053
|
0.703
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr8_+_19841120
|
0.699
|
|
LPL
|
lipoprotein lipase
|
|
chr17_-_44047299
|
0.695
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr15_-_38387357
|
0.692
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr7_-_100021658
|
0.690
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr7_-_27206249
|
0.684
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr2_-_238813365
|
0.683
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr12_-_8656705
|
0.675
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr1_+_154386358
|
0.670
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr3_+_129124591
|
0.670
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr9_+_94766379
|
0.669
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr15_-_81037554
|
0.669
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr14_-_106241445
|
0.669
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr16_+_28850760
|
0.668
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr13_-_69580459
|
0.662
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr6_+_34313007
|
0.662
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_-_193928038
|
0.653
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr6_-_31668476
|
0.647
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr12_-_68291208
|
0.643
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr6_+_34312641
|
0.641
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr11_-_113935789
|
0.640
|
NM_152315
|
FAM55A
|
family with sequence similarity 55, member A
|
|
chr15_-_38387410
|
0.636
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chrX_+_37524190
|
0.629
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr2_-_238813284
|
0.629
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr11_+_527521
|
0.623
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr17_-_34635446
|
0.615
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr1_+_165564904
|
0.612
|
NM_001198783
|
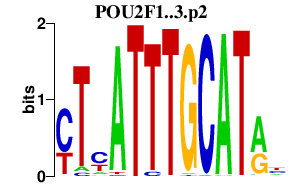
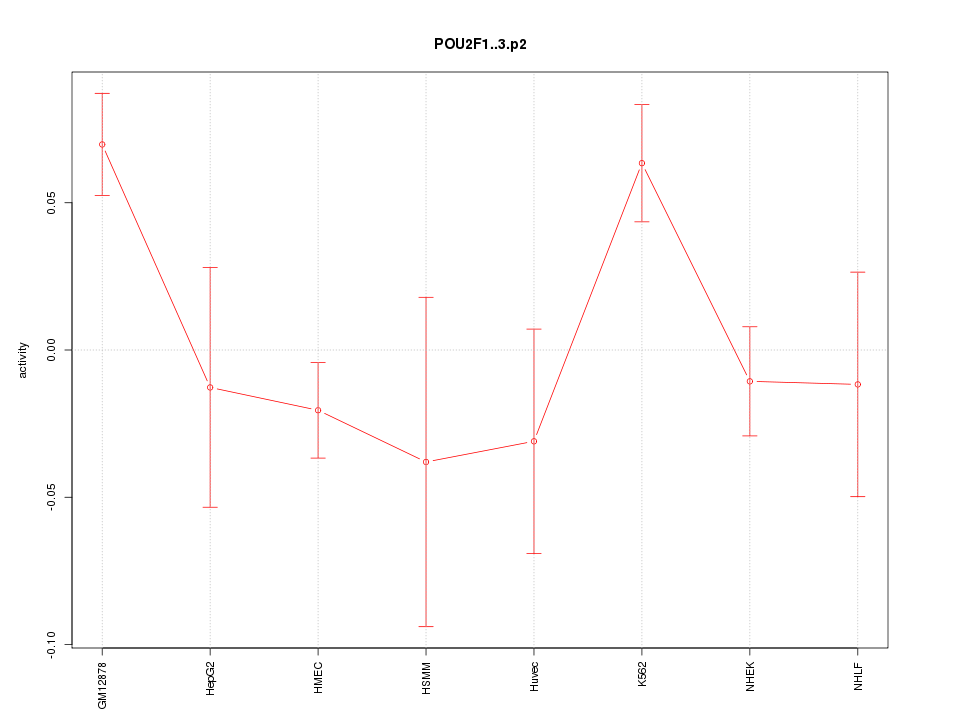
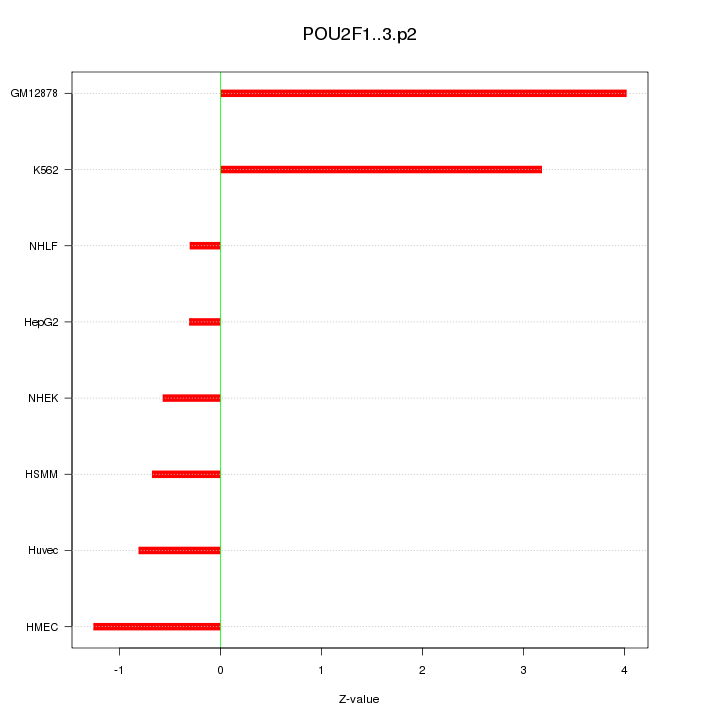
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_-_84521398
|
0.610
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr18_-_22696532
|
0.609
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr18_+_74841262
|
0.608
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr7_+_37746520
|
0.593
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr1_-_203579932
|
0.592
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr17_-_33179118
|
0.591
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_204797079
|
0.589
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr6_+_12825022
|
0.574
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr6_-_99904251
|
0.573
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr1_+_148125090
|
0.573
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr17_-_53849888
|
0.569
|
|
RNF43
|
ring finger protein 43
|
|
chr13_-_52320774
|
0.564
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr17_-_4792542
|
0.560
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr17_-_36850978
|
0.559
|
NM_006771
|
KRT38
|
keratin 38
|
|
chr8_-_72437020
|
0.556
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr17_+_60563917
|
0.555
|
NM_001081955
NM_001165933
NM_003835
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr3_+_120799411
|
0.549
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chrX_+_153767803
|
0.548
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr7_-_44088608
|
0.540
|
|
POLM
|
polymerase (DNA directed), mu
|
|
chr5_+_2805261
|
0.536
|
NM_178569
|
C5orf38
|
chromosome 5 open reading frame 38
|
|
chr6_+_34312625
|
0.534
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_204797120
|
0.528
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr10_+_68355797
|
0.522
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr8_-_25371663
|
0.521
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr3_-_167037861
|
0.519
|
|
BCHE
|
butyrylcholinesterase
|
|
chr11_-_117552545
|
0.517
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr12_+_120548831
|
0.513
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr1_-_194844061
|
0.512
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr19_-_17377383
|
0.503
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr22_+_21384862
|
0.503
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr14_+_22422270
|
0.502
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr22_-_14829803
|
0.502
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chr9_+_128716867
|
0.499
|
NM_001190729
NM_001190730
NM_014636
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr15_-_56145140
|
0.498
|
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr3_+_63613383
|
0.497
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr14_-_105681377
|
0.494
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_+_63829616
|
0.488
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr16_-_54466709
|
0.483
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr2_-_51112743
|
0.481
|
|
NRXN1
|
neurexin 1
|
|
chr6_+_34312633
|
0.481
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_+_21116283
|
0.477
|
|
|
|
|
chr6_+_34312554
|
0.476
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr18_-_59462469
|
0.475
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr3_-_167037946
|
0.473
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr2_-_104834344
|
0.466
|
|
LOC100506421
|
hypothetical LOC100506421
|