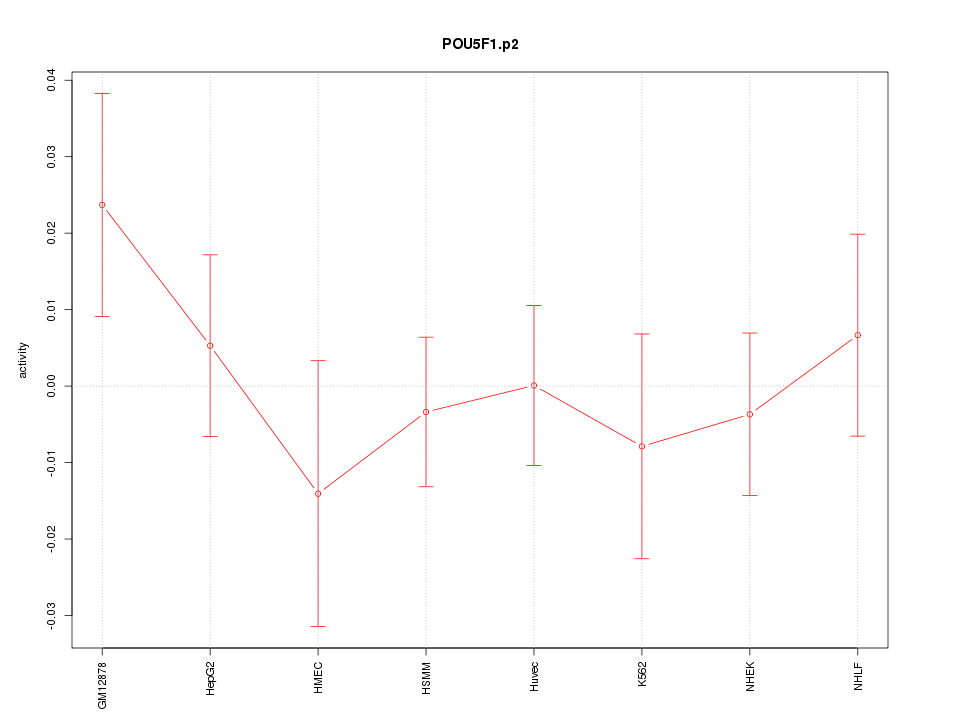
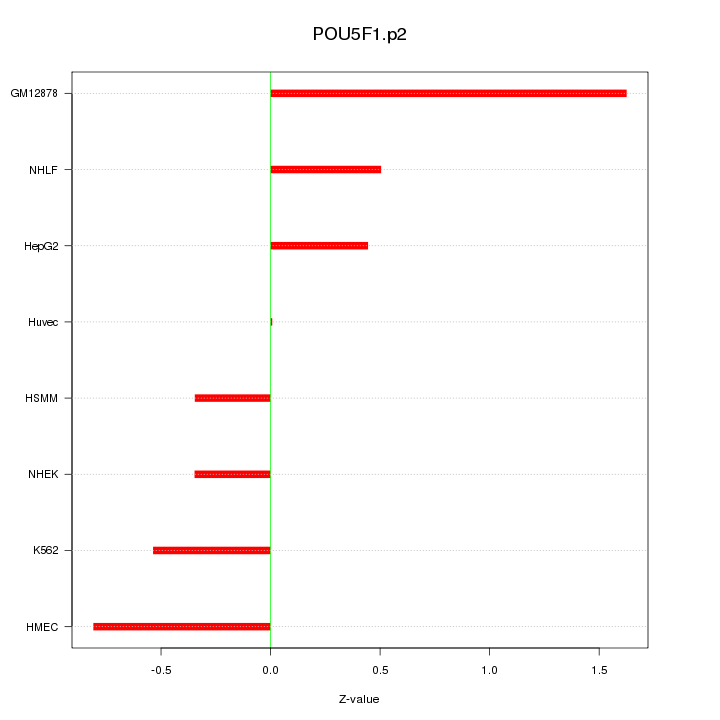
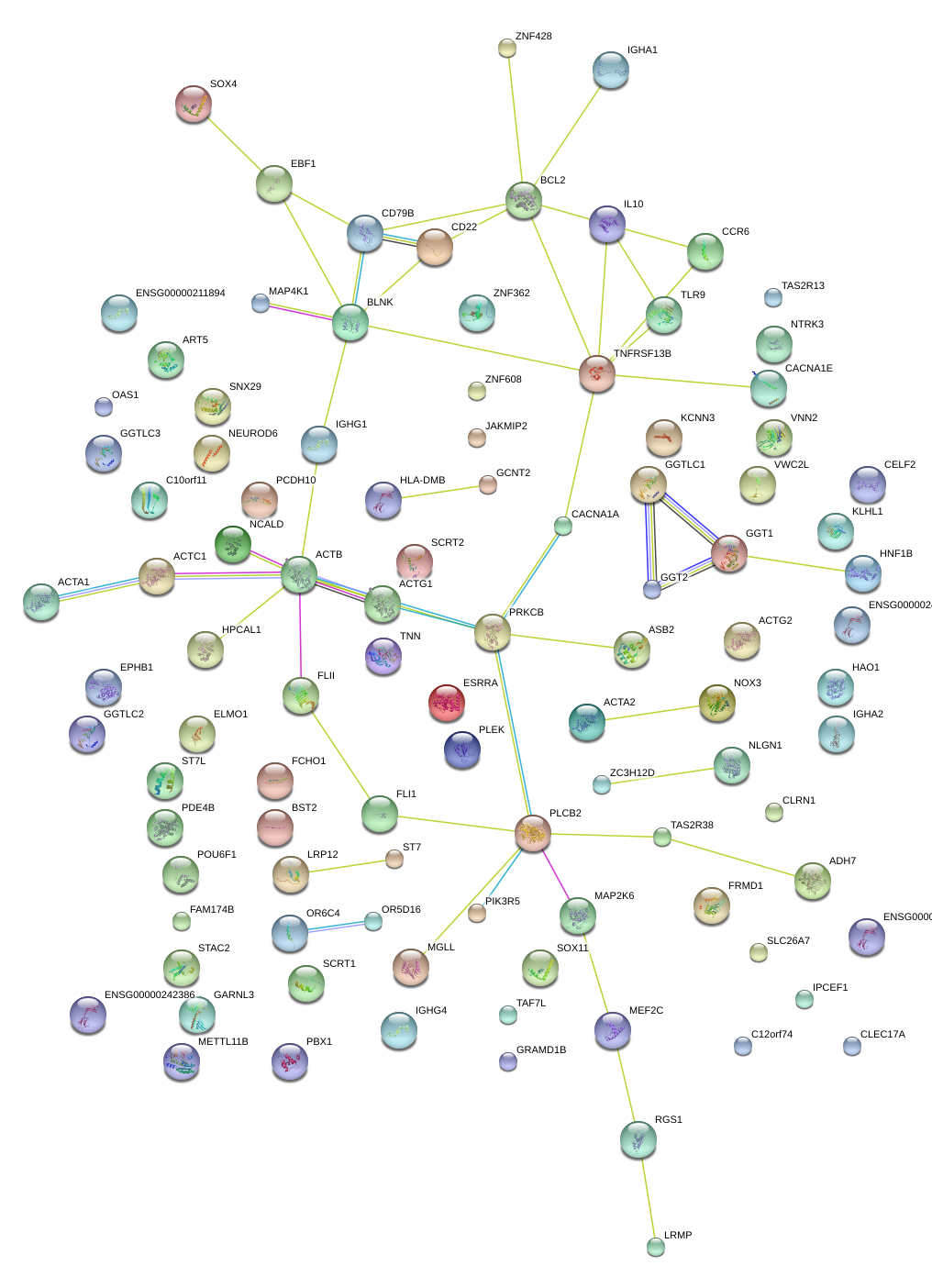
|
chr17_+_64922420
|
4.787
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr10_+_11246934
|
1.380
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_153098811
|
1.359
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_-_36991189
|
1.333
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_-_8756524
|
1.267
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr1_+_190552744
|
1.160
|
NM_001039152
|
RGS21
|
regulator of G-protein signaling 21
|
|
chr21_-_31123797
|
1.022
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr20_-_7869068
|
0.930
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr10_-_98021144
|
0.902
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr3_-_152173475
|
0.898
|
NM_001195794
NM_174878
|
CLRN1
|
clarin 1
|
|
chr9_+_129066576
|
0.885
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr16_+_23754772
|
0.837
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr6_-_154692906
|
0.821
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr8_+_92330691
|
0.811
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr5_-_147142249
|
0.800
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr22_+_21318779
|
0.777
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr6_-_33016561
|
0.776
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr15_-_86600658
|
0.760
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr1_+_66230977
|
0.748
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr11_+_122901543
|
0.741
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr19_+_14554895
|
0.736
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr19_-_43800374
|
0.732
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr14_-_93512818
|
0.692
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr2_+_214984705
|
0.690
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr10_+_11247059
|
0.688
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr19_-_48815853
|
0.683
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr5_-_88155360
|
0.679
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_155818728
|
0.673
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr12_+_111829121
|
0.649
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr12_+_91620749
|
0.645
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr11_+_55362802
|
0.639
|
NM_001005496
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16
|
|
chr16_+_12053555
|
0.638
|
NM_001080530
|
SNX29
|
sorting nexin 29
|
|
chr17_-_33179118
|
0.634
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr19_+_17719504
|
0.625
|
NM_001161357
NM_001161358
|
FCHO1
|
FCH domain only 1
|
|
chr11_+_122901689
|
0.609
|
NM_020716
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr1_+_33494745
|
0.606
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr11_+_128068982
|
0.587
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_+_190811479
|
0.579
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr17_-_16816113
|
0.575
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr6_+_167456230
|
0.575
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr12_+_54231249
|
0.573
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chr1_+_162795328
|
0.569
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr4_+_134289893
|
0.567
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr7_+_116447527
|
0.564
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr15_-_91078307
|
0.562
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr20_-_604822
|
0.562
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr6_-_149847809
|
0.561
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr2_+_73973600
|
0.555
|
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr11_-_3620060
|
0.548
|
NM_053017
NM_001079536
|
ART5
|
ADP-ribosyltransferase 5
|
|
chr6_-_149847718
|
0.537
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr7_-_31347032
|
0.531
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr18_+_52965290
|
0.529
|
|
BOD1P
|
biorientation of chromosomes in cell division 1 pseudogene
|
|
chr17_-_59363352
|
0.526
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr3_+_135996734
|
0.518
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr17_-_33178987
|
0.516
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr14_-_93512647
|
0.516
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr1_+_168381811
|
0.513
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr18_-_59137488
|
0.510
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr19_+_40511909
|
0.504
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chr4_-_100575313
|
0.502
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr3_-_129023850
|
0.498
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr1_+_162795538
|
0.498
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr14_-_106084519
|
0.497
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_124108672
|
0.496
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr10_+_77212524
|
0.492
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr1_+_179719338
|
0.482
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr6_+_10663934
|
0.482
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr3_-_52235218
|
0.475
|
NM_017442
|
TLR9
|
toll-like receptor 9
|
|
chr3_+_135996990
|
0.474
|
|
EPHB1
|
EPH receptor B1
|
|
chr8_-_102872354
|
0.469
|
|
NCALD
|
neurocalcin delta
|
|
chr22_+_21065134
|
0.465
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr15_-_38387410
|
0.463
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr19_-_17377383
|
0.457
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr17_-_34635446
|
0.457
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr12_+_25096755
|
0.455
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr2_+_68445821
|
0.455
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr2_+_5750229
|
0.454
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr3_+_174598937
|
0.453
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr16_+_23754860
|
0.450
|
|
PRKCB
|
protein kinase C, beta
|
|
chr12_-_49897743
|
0.447
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr6_+_10693964
|
0.445
|
NM_145655
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr6_-_133120587
|
0.442
|
NM_004665
|
VNN2
|
vanin 2
|
|
chr6_-_168222645
|
0.442
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr13_-_69580459
|
0.437
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr12_-_10953427
|
0.436
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr7_-_141320041
|
0.436
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr1_+_173303616
|
0.435
|
NM_022093
|
TNN
|
tenascin N
|
|
chr1_-_205012348
|
0.433
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr11_+_63829616
|
0.429
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr5_-_158459006
|
0.427
|
|
EBF1
|
early B-cell factor 1
|
|
chr19_-_48815572
|
0.426
|
|
ZNF428
|
zinc finger protein 428
|
|
chr5_-_175897430
|
0.426
|
|
RNF44
|
ring finger protein 44
|
|
chr7_-_126670804
|
0.421
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr5_-_111119846
|
0.421
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_-_56342097
|
0.420
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr13_-_27572646
|
0.418
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr2_+_17861266
|
0.417
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr12_-_16321885
|
0.410
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr17_-_39094787
|
0.410
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr16_+_23754822
|
0.409
|
|
PRKCB
|
protein kinase C, beta
|
|
chr20_+_17155680
|
0.408
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr17_-_38087366
|
0.408
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr16_+_31273969
|
0.407
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr3_+_51837608
|
0.403
|
NM_001085479
|
IQCF3
|
IQ motif containing F3
|
|
chr4_-_41579325
|
0.400
|
|
|
|
|
chrX_-_84521398
|
0.399
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr1_+_148125090
|
0.399
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr18_-_54447168
|
0.399
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr12_-_7136147
|
0.391
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr16_+_31274030
|
0.390
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr20_+_17155669
|
0.390
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr1_+_5975345
|
0.388
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr15_-_78050515
|
0.388
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr5_-_156322784
|
0.384
|
NM_001146726
NM_138379
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4
|
|
chr6_-_32665409
|
0.383
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr2_-_50428395
|
0.381
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr10_+_124897627
|
0.381
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr3_+_99733432
|
0.380
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr10_+_68355797
|
0.379
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr12_+_109956210
|
0.377
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chrX_-_80344096
|
0.377
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr12_+_25096472
|
0.374
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr7_+_50318801
|
0.372
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr10_+_106103503
|
0.371
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr7_+_149842850
|
0.371
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr8_-_72431274
|
0.366
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr5_-_160211623
|
0.366
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr22_-_35938298
|
0.361
|
NM_001051
|
SSTR3
|
somatostatin receptor 3
|
|
chr2_+_189547358
|
0.361
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_5975286
|
0.360
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr20_-_43693320
|
0.359
|
NM_147198
|
WFDC9
|
WAP four-disulfide core domain 9
|
|
chr7_-_100021658
|
0.358
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr1_+_154120170
|
0.358
|
|
SYT11
|
synaptotagmin XI
|
|
chr17_-_60988077
|
0.357
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr7_+_73826244
|
0.355
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr3_-_57209319
|
0.354
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr6_-_42527760
|
0.353
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chrX_+_16051344
|
0.352
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr11_+_128069183
|
0.351
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr6_-_108252200
|
0.350
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr9_+_94766379
|
0.348
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr12_-_120502483
|
0.348
|
NM_001005366
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr5_-_88215034
|
0.347
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_+_116447499
|
0.346
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr2_-_157890400
|
0.345
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr17_+_60563917
|
0.344
|
NM_001081955
NM_001165933
NM_003835
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr12_+_111829191
|
0.343
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr6_+_26325124
|
0.343
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr16_+_23754997
|
0.342
|
|
PRKCB
|
protein kinase C, beta
|
|
chr22_+_30072739
|
0.338
|
|
FLJ20464
|
hypothetical protein FLJ20464
|
|
chr3_+_129124591
|
0.337
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr3_+_141879555
|
0.336
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr2_+_73973640
|
0.336
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr14_+_31868229
|
0.335
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr17_-_44249379
|
0.335
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr17_+_60563963
|
0.335
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr7_-_3049872
|
0.334
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr3_-_193928038
|
0.333
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr12_-_103846186
|
0.333
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr8_-_72437020
|
0.333
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_-_27206249
|
0.332
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr6_+_29187565
|
0.331
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr14_+_78815406
|
0.330
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr8_+_144711619
|
0.329
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr22_-_30072329
|
0.328
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr5_-_66528330
|
0.327
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr12_-_7709734
|
0.326
|
NM_001644
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
|
|
chr22_+_20715340
|
0.326
|
|
|
|
|
chr12_-_101398429
|
0.326
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr7_-_100021689
|
0.323
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr6_+_29902734
|
0.322
|
NM_002127
|
HLA-G
|
major histocompatibility complex, class I, G
|
|
chr10_+_120957084
|
0.322
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr9_-_35069751
|
0.322
|
NM_004629
|
FANCG
|
Fanconi anemia, complementation group G
|
|
chr11_-_10630408
|
0.321
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr3_+_185300660
|
0.320
|
NM_182589
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3, family member E
|
|
chr14_+_99555471
|
0.320
|
|
EVL
|
Enah/Vasp-like
|
|
chr1_+_165564904
|
0.319
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr11_+_128069203
|
0.318
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr12_-_51086330
|
0.318
|
NM_033033
|
KRT82
|
keratin 82
|
|
chr18_+_40513889
|
0.318
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr17_+_9670105
|
0.315
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr11_+_4522993
|
0.315
|
NM_001004137
|
OR52M1
|
olfactory receptor, family 52, subfamily M, member 1
|
|
chr1_-_158948176
|
0.314
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr22_-_30072217
|
0.313
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr1_+_100957883
|
0.313
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr12_-_101398470
|
0.313
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_70639382
|
0.310
|
NM_014595
|
NT5C
|
5', 3'-nucleotidase, cytosolic
|
|
chr14_-_105851580
|
0.310
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_-_84521320
|
0.310
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr17_-_23151601
|
0.309
|
NM_000625
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr14_-_88948486
|
0.309
|
|
FOXN3
|
forkhead box N3
|
|
chr8_+_11388918
|
0.308
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr6_-_11340886
|
0.308
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_+_135558744
|
0.303
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr15_+_58083712
|
0.302
|
NM_012182
|
FOXB1
|
forkhead box B1
|
|
chr14_-_106184988
|
0.302
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_+_100021862
|
0.300
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr1_+_218930250
|
0.300
|
NM_024709
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chrX_+_28515479
|
0.298
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr8_+_19840861
|
0.297
|
NM_000237
|
LPL
|
lipoprotein lipase
|