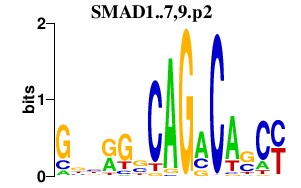
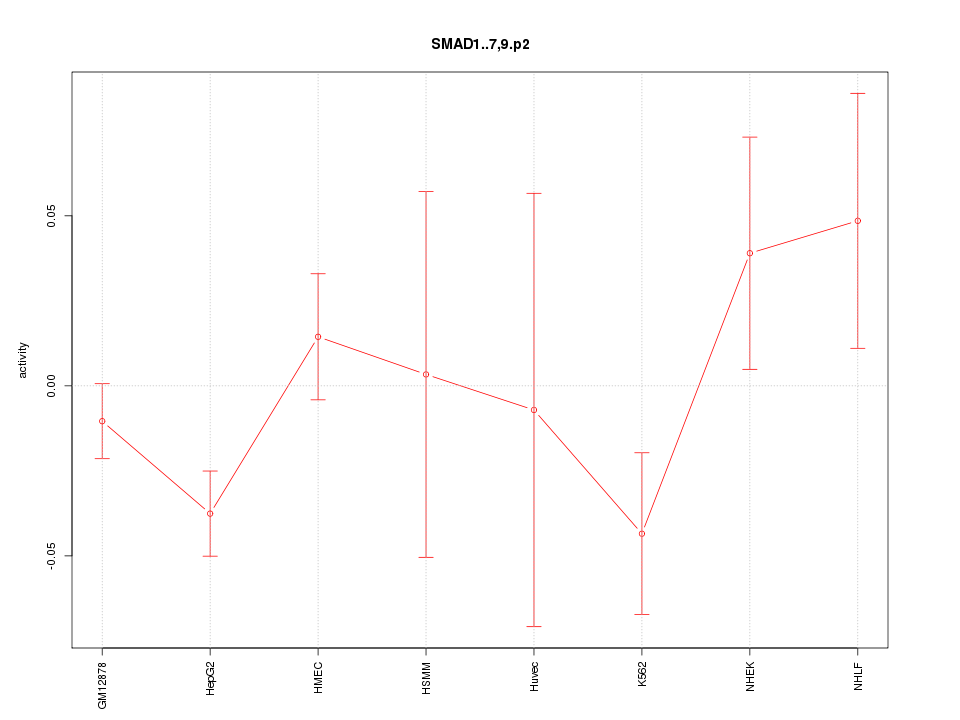
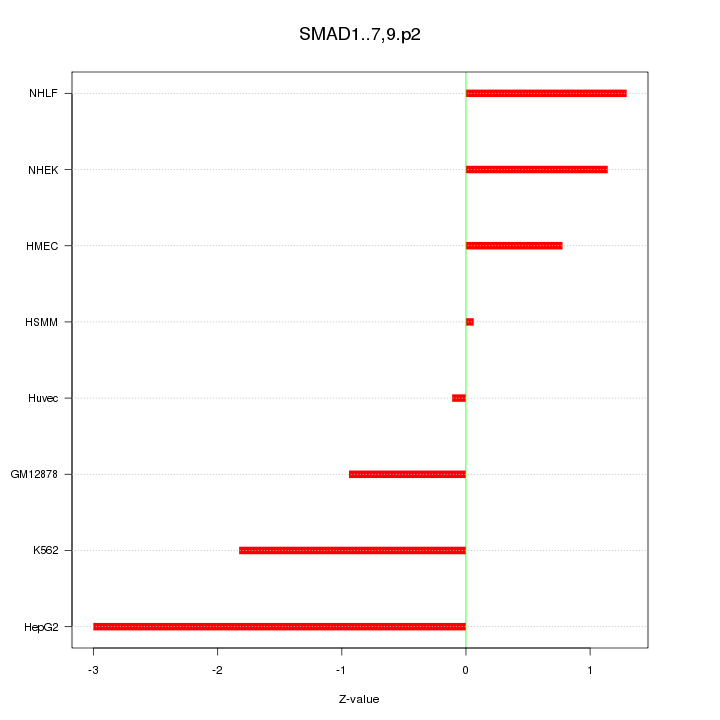
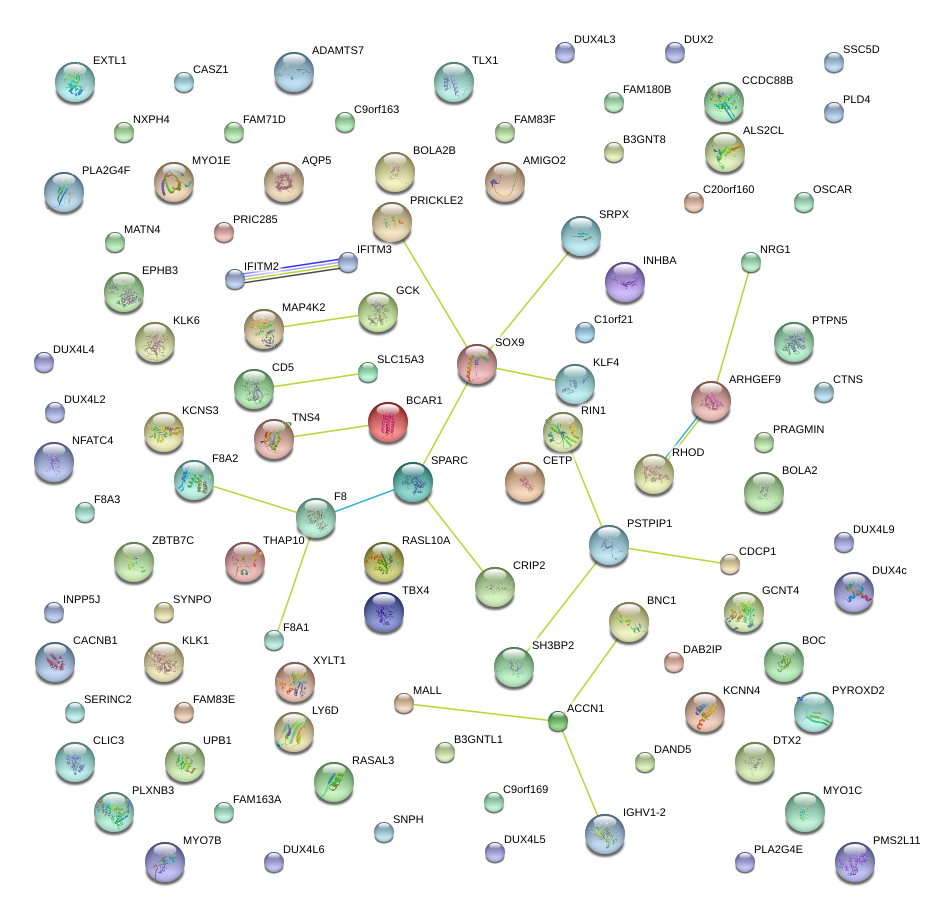
|
chr11_-_65860450
|
4.226
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr11_-_18769703
|
2.774
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chrX_+_152682772
|
2.716
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr12_+_55896844
|
2.583
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr8_+_32623792
|
2.571
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr11_+_60626505
|
2.342
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr10_+_135333667
|
2.266
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr15_-_40236061
|
2.153
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr11_+_66580884
|
2.119
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr10_+_135343649
|
2.089
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr4_+_191239247
|
2.004
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr20_+_1194959
|
1.993
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr14_+_104462260
|
1.948
|
|
PLD4
|
phospholipase D family, member 4
|
|
chr20_+_30061905
|
1.938
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr20_-_43370333
|
1.929
|
NM_003833
NM_030590
NM_030592
|
MATN4
|
matrilin 4
|
|
chr15_-_76890626
|
1.902
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr1_+_26220852
|
1.837
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr3_-_64186151
|
1.833
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr8_-_143864933
|
1.763
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr2_-_110231431
|
1.743
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr4_+_191226073
|
1.740
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr12_+_56406296
|
1.726
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr11_+_47564805
|
1.716
|
NM_001164379
|
FAM180B
|
family with sequence similarity 180, member B
|
|
chr19_-_59295882
|
1.714
|
NM_130771
NM_133168
NM_133169
NM_206818
|
OSCAR
|
osteoclast associated, immunoglobulin-like receptor
|
|
chr1_+_182622850
|
1.713
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr19_-_46626474
|
1.704
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr3_+_114414064
|
1.689
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr7_-_44195405
|
1.688
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr15_-_81744469
|
1.669
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr3_+_185762233
|
1.665
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr10_-_100164877
|
1.630
|
NM_032709
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2
|
|
chr4_+_191242540
|
1.622
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191232653
|
1.615
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr11_-_310775
|
1.596
|
NM_021034
|
IFITM3
|
interferon induced transmembrane protein 3 (1-8U)
|
|
chr17_-_1342700
|
1.590
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr1_+_26220895
|
1.589
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr2_+_128009782
|
1.576
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr4_+_191235959
|
1.551
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr9_+_123369219
|
1.549
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr1_+_182622751
|
1.547
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr18_-_43821491
|
1.541
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr5_-_151046638
|
1.498
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr7_-_41709191
|
1.492
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr9_-_109291130
|
1.489
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_177978920
|
1.466
|
NM_173509
|
FAM163A
|
family with sequence similarity 163, member A
|
|
chr19_+_12941431
|
1.463
|
NM_152654
|
DAND5
|
DAN domain family, member 5
|
|
chr16_-_29373785
|
1.445
|
NM_001031827
NM_001039182
|
BOLA2
BOLA2B
|
bolA homolog 2 (E. coli)
bolA homolog 2B (E. coli)
|
|
chr11_-_310925
|
1.436
|
|
IFITM3
|
interferon induced transmembrane protein 3 (1-8U)
|
|
chr9_+_139239464
|
1.417
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr11_+_63864263
|
1.412
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr10_+_102881050
|
1.409
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chrX_-_154341451
|
1.390
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr12_-_45759691
|
1.354
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr22_-_28041650
|
1.353
|
NM_001007279
NM_006477
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr9_-_139010805
|
1.340
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr5_-_74362479
|
1.330
|
NM_016591
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chr19_-_15436333
|
1.325
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr15_+_75074483
|
1.325
|
NM_003978
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr20_-_61669550
|
1.314
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr14_+_23908141
|
1.313
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr17_+_67628749
|
1.309
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr19_+_60691671
|
1.300
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr7_+_75947762
|
1.278
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr17_+_56884560
|
1.271
|
|
TBX4
|
T-box 4
|
|
chr10_-_102880892
|
1.265
|
NM_001085398
|
TLX1NB
|
TLX1 neighbor
|
|
chr2_+_17923404
|
1.263
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr17_+_3486753
|
1.251
|
|
CTNS
|
cystinosis, nephropathic
|
|
chr16_-_73830025
|
1.250
|
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr11_+_66580861
|
1.227
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr3_-_46693902
|
1.214
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr17_-_34607257
|
1.211
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr1_-_10779291
|
1.209
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr22_-_28041474
|
1.202
|
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr1_+_31659246
|
1.200
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr16_+_55553262
|
1.191
|
NM_000078
|
CETP
|
cholesteryl ester transfer protein, plasma
|
|
chr5_-_151046680
|
1.190
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr9_+_138497767
|
1.190
|
NM_152571
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chr5_+_149960820
|
1.189
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr4_+_191229360
|
1.187
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_-_53808505
|
1.184
|
NM_017708
|
FAM83E
|
family with sequence similarity 83, member E
|
|
chr22_+_29848923
|
1.169
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr22_+_23221114
|
1.168
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr3_-_45162901
|
1.153
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr22_+_38720881
|
1.143
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr12_+_48641545
|
1.138
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr17_-_35911340
|
1.138
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr11_-_60475558
|
1.135
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr16_-_17472238
|
1.135
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr14_+_105012106
|
1.130
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr4_+_2783764
|
1.119
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr19_-_48976852
|
1.116
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr14_+_66725862
|
1.104
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr15_-_68971665
|
1.102
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr15_-_40089736
|
1.097
|
NM_001080490
|
PLA2G4E
|
phospholipase A2, group IVE
|
|
chrX_-_37916138
|
1.095
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chrX_-_62891730
|
1.093
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr3_-_45162847
|
1.093
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr15_+_75074743
|
1.092
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr8_-_8276598
|
1.090
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr19_-_56164628
|
1.084
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr5_+_72452131
|
1.070
|
NM_001161342
NM_173490
|
TMEM171
|
transmembrane protein 171
|
|
chr19_+_40299058
|
1.068
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr3_+_89239363
|
1.065
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr17_+_63713171
|
1.060
|
|
|
|
|
chr11_+_129823667
|
1.055
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr1_+_4614528
|
1.054
|
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr13_-_39075304
|
1.053
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr20_-_57948653
|
1.048
|
NM_006242
|
PPP1R3D
|
protein phosphatase 1, regulatory (inhibitor) subunit 3D
|
|
chr21_-_41479035
|
1.048
|
NM_182832
|
PLAC4
|
placenta-specific 4
|
|
chr18_+_40513889
|
1.048
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr1_-_27212035
|
1.044
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr19_+_43572679
|
1.044
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr1_+_8928508
|
1.038
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chrX_+_133952633
|
1.029
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr19_-_17275083
|
1.028
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr15_+_78774664
|
1.020
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr4_+_72271218
|
1.015
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_+_191245840
|
1.013
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_+_199426575
|
1.010
|
NM_001164586
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
|
chr5_+_170779664
|
1.009
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr16_+_87232501
|
1.005
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr1_+_181422016
|
1.004
|
|
LAMC2
|
laminin, gamma 2
|
|
chr8_+_22493928
|
1.001
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr3_-_197106828
|
0.995
|
NM_001010938
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr19_+_60691733
|
0.989
|
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr1_-_158091760
|
0.988
|
NM_001134233
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr19_-_48977247
|
0.987
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr16_+_56219802
|
0.987
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr4_+_36923084
|
0.986
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr1_+_182622987
|
0.985
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr21_+_38550739
|
0.984
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr5_-_146813343
|
0.982
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr12_+_45759634
|
0.978
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr12_+_2032964
|
0.970
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr11_+_64114857
|
0.968
|
NM_144585
NM_153378
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12
|
|
chr3_-_197103848
|
0.967
|
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr19_-_48133089
|
0.966
|
NM_002783
|
PSG7
|
pregnancy specific beta-1-glycoprotein 7 (gene/pseudogene)
|
|
chr8_-_144726064
|
0.966
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr10_-_128067062
|
0.960
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr3_-_48576204
|
0.955
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr21_+_46370831
|
0.946
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_-_34491878
|
0.943
|
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr19_+_45807909
|
0.941
|
|
|
|
|
chr17_+_74531820
|
0.940
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_-_52667056
|
0.933
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr1_+_181421984
|
0.933
|
|
LAMC2
|
laminin, gamma 2
|
|
chr17_-_7238546
|
0.930
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr19_-_41034734
|
0.924
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr6_+_26609427
|
0.924
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr9_-_139455611
|
0.920
|
NM_001033113
NM_198585
|
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8
|
|
chrX_-_132376660
|
0.919
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr1_-_41722883
|
0.918
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr14_+_64086366
|
0.917
|
NM_172365
|
C14orf50
|
chromosome 14 open reading frame 50
|
|
chr5_+_170779264
|
0.917
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr11_+_124114985
|
0.916
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr3_-_129024665
|
0.912
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr19_+_43446937
|
0.909
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr10_-_128067052
|
0.900
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_+_181421796
|
0.900
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr19_-_53912025
|
0.898
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr5_-_149515502
|
0.898
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr21_+_44844924
|
0.894
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr1_+_15964045
|
0.893
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr1_-_27211913
|
0.893
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr1_-_112333192
|
0.892
|
NM_004980
NM_172198
|
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3
|
|
chr8_-_144770764
|
0.891
|
|
TSTA3
|
tissue specific transplantation antigen P35B
|
|
chr9_-_109291575
|
0.890
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr5_-_127447197
|
0.890
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr5_-_156468614
|
0.885
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chrX_-_62891667
|
0.885
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr11_+_122258600
|
0.884
|
NM_024806
NM_199124
|
C11orf63
|
chromosome 11 open reading frame 63
|
|
chr19_-_6230797
|
0.881
|
|
|
|
|
chr9_-_138387926
|
0.880
|
NM_052813
NM_052814
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chrX_+_102749489
|
0.874
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr1_+_199884152
|
0.874
|
|
NAV1
|
neuron navigator 1
|
|
chr22_+_36286350
|
0.874
|
NM_152243
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr3_-_45162783
|
0.873
|
|
|
|
|
chr17_+_74531873
|
0.867
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr8_-_134378630
|
0.860
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr22_+_42137955
|
0.858
|
NM_001044370
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr19_+_50663089
|
0.857
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr1_-_145712107
|
0.854
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr22_+_29333069
|
0.852
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr21_+_44856423
|
0.850
|
NM_198695
|
KRTAP10-8
|
keratin associated protein 10-8
|
|
chr4_-_8211335
|
0.847
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr20_-_61676003
|
0.846
|
NM_001037335
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr17_-_59403932
|
0.845
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chrX_+_149280486
|
0.845
|
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr18_-_55091596
|
0.842
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr9_-_21964825
|
0.841
|
NM_058197
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr3_+_11171213
|
0.838
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr1_+_155130146
|
0.837
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr11_-_110888273
|
0.836
|
NM_017589
|
BTG4
|
B-cell translocation gene 4
|
|
chr21_+_38550533
|
0.836
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr20_-_17459973
|
0.835
|
NM_001195
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr9_-_21965131
|
0.834
|
NM_000077
NM_001195132
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chrX_+_106956115
|
0.831
|
|
MID2
|
midline 2
|
|
chr3_-_140246423
|
0.829
|
NM_001134657
|
PRR23C
|
proline rich 23C
|
|
chr20_+_52525538
|
0.829
|
NM_018431
|
DOK5
|
docking protein 5
|
|
chr1_+_210864411
|
0.828
|
NM_153606
|
FAM71A
|
family with sequence similarity 71, member A
|