|
chr5_-_139761182
|
1.464
|
|
|
|
|
chr11_+_13646727
|
1.385
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_+_13646806
|
1.161
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_-_57173823
|
1.074
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr18_-_21186107
|
1.064
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr8_-_9046460
|
1.035
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr14_-_22458134
|
1.025
|
|
RBM23
|
RNA binding motif protein 23
|
|
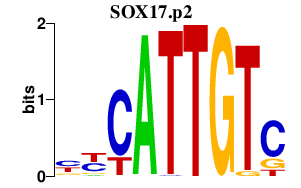
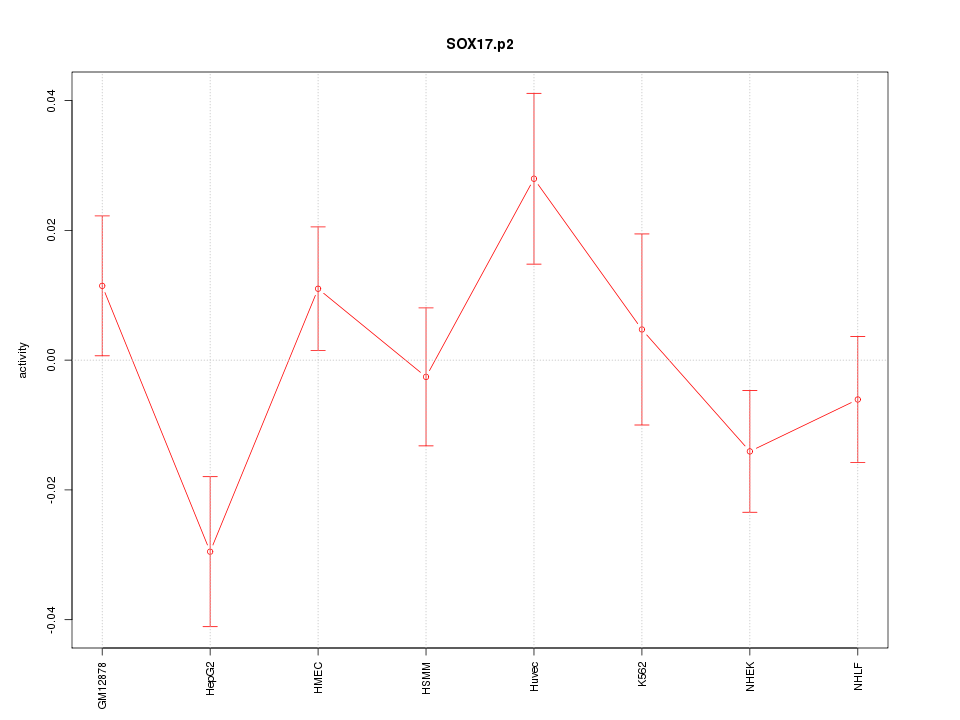
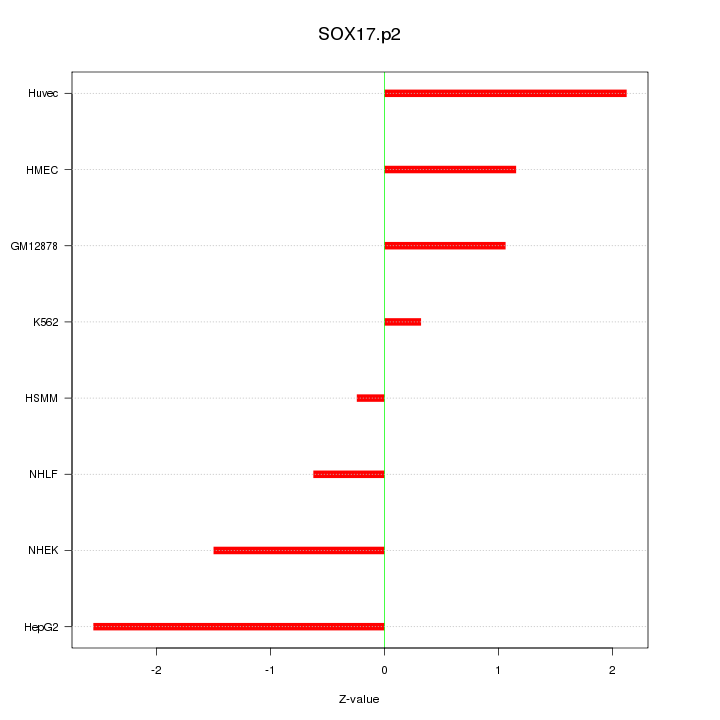
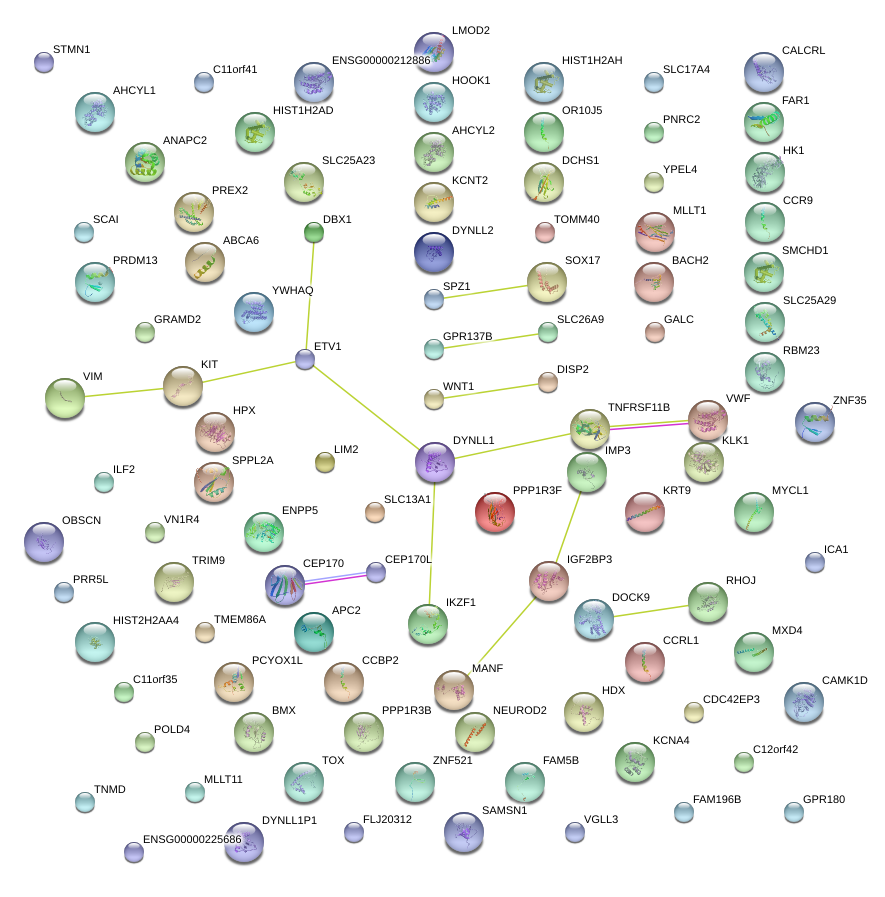
chr8_+_55533047
|
1.010
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chrX_+_49013243
|
0.994
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr2_-_37752272
|
0.987
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_-_35017691
|
0.972
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chrX_-_83644054
|
0.934
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr11_+_13646839
|
0.865
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr14_-_22458199
|
0.844
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr19_-_56582997
|
0.840
|
NM_001161748
NM_030657
|
LIM2
|
lens intrinsic membrane protein 2, 19kDa
|
|
chr2_-_188021117
|
0.835
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr6_-_72187168
|
0.824
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr6_+_25862905
|
0.818
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chrX_+_15428820
|
0.811
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr1_+_175406685
|
0.802
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr5_+_79651545
|
0.799
|
NM_032567
|
SPZ1
|
spermatogenic leucine zipper 1
|
|
chr14_-_87529600
|
0.785
|
NM_000153
NM_001037525
|
GALC
|
galactosylceramidase
|
|
chr19_-_6230927
|
0.768
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr14_-_99842519
|
0.750
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr1_-_151910080
|
0.747
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr4_-_2233504
|
0.744
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr7_+_50314801
|
0.728
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_-_26105477
|
0.723
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr15_-_70277163
|
0.719
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr7_-_122627235
|
0.702
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr11_-_66877519
|
0.701
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr9_-_126945536
|
0.691
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr3_+_51397756
|
0.688
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr15_-_73719546
|
0.670
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr15_+_38437703
|
0.659
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr14_-_22458220
|
0.648
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr13_+_94052022
|
0.647
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr21_-_14840506
|
0.639
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr14_+_62740878
|
0.634
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr17_-_59817722
|
0.631
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr3_+_44665220
|
0.630
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr1_-_241485105
|
0.629
|
|
CEP170
|
centrosomal protein 170kDa
|
|
chr10_+_17311282
|
0.628
|
|
VIM
|
vimentin
|
|
chr12_+_119418294
|
0.626
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr2_-_37752731
|
0.621
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr8_-_60194099
|
0.620
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr1_-_157772420
|
0.610
|
NM_001004469
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5
|
|
chr4_+_55218663
|
0.609
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr10_+_70748581
|
0.602
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr11_-_550706
|
0.601
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr6_+_100161370
|
0.601
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr3_-_87122936
|
0.595
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr7_-_8268666
|
0.584
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr11_-_6633427
|
0.582
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr19_-_58462729
|
0.572
|
NM_173857
|
VN1R4
|
vomeronasal 1 receptor 4
|
|
chr12_-_102413875
|
0.572
|
NM_198521
NM_001099336
|
C12orf42
|
chromosome 12 open reading frame 42
|
|
chr5_-_169340321
|
0.571
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr6_-_91063301
|
0.568
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_-_6103945
|
0.564
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr3_+_45902999
|
0.563
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr5_+_148717762
|
0.563
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr6_+_27222839
|
0.562
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr1_-_26105230
|
0.555
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr17_-_64649608
|
0.549
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr2_-_9688450
|
0.542
|
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr6_-_91063181
|
0.538
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr4_-_2233606
|
0.529
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr19_+_50086362
|
0.528
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr5_-_139760838
|
0.525
|
|
|
|
|
chr1_-_40140126
|
0.520
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr17_-_59817456
|
0.519
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr5_+_148717806
|
0.518
|
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr8_+_69027143
|
0.517
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr8_-_120033240
|
0.517
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr1_+_24158898
|
0.516
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr10_+_12431477
|
0.513
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr11_+_33520348
|
0.512
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr11_+_13646822
|
0.509
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_+_36274300
|
0.507
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr1_-_204179169
|
0.504
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr7_+_128802719
|
0.504
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr18_-_21185968
|
0.503
|
|
ZNF521
|
zinc finger protein 521
|
|
chrX_+_99726445
|
0.503
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr14_-_87529222
|
0.500
|
|
GALC
|
galactosylceramidase
|
|
chr11_-_6418705
|
0.500
|
NM_000613
|
HPX
|
hemopexin
|
|
chr13_-_98428244
|
0.499
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr1_+_207668787
|
0.498
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr14_-_50631407
|
0.497
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr12_+_47658502
|
0.495
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr3_+_51397731
|
0.495
|
NM_006010
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr11_+_18676913
|
0.492
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr2_-_9688552
|
0.492
|
NM_006826
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr7_-_13995811
|
0.489
|
|
ETV1
|
ets variant 1
|
|
chr14_-_87529212
|
0.489
|
|
GALC
|
galactosylceramidase
|
|
chr1_+_234372454
|
0.488
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr1_+_149299504
|
0.488
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_-_46246543
|
0.485
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr1_-_194844061
|
0.483
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr7_+_123083096
|
0.481
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr19_+_1401208
|
0.479
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr9_-_35804997
|
0.479
|
|
HINT2
|
histidine triad nucleotide binding protein 2
|
|
chrX_+_136475979
|
0.475
|
NM_003413
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr19_+_13767207
|
0.474
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr10_+_17311962
|
0.474
|
|
VIM
|
vimentin
|
|
chr11_-_4585947
|
0.473
|
NM_018073
|
TRIM68
|
tripartite motif containing 68
|
|
chr8_-_22606653
|
0.472
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_-_234096793
|
0.471
|
|
|
|
|
chr15_-_39973566
|
0.469
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr12_+_8557402
|
0.468
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr18_+_29412538
|
0.466
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr2_-_144991386
|
0.466
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_71747867
|
0.465
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr22_-_30981317
|
0.462
|
NM_014227
|
SLC5A4
|
solute carrier family 5 (low affinity glucose cotransporter), member 4
|
|
chr6_-_20320607
|
0.460
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr1_+_43539239
|
0.460
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr1_+_199975674
|
0.457
|
|
NAV1
|
neuron navigator 1
|
|
chr11_+_128068982
|
0.452
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_-_170347019
|
0.451
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_59010797
|
0.450
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr1_+_149299530
|
0.443
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_149299490
|
0.442
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr19_+_50086357
|
0.441
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chrX_+_82649924
|
0.438
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr5_+_40715778
|
0.435
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr20_-_55718982
|
0.433
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr5_-_41107161
|
0.431
|
NM_173489
|
HEATR7B2
|
HEAT repeat family member 7B2
|
|
chr14_+_85066207
|
0.431
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr5_-_157011949
|
0.431
|
NM_007017
NM_178424
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr19_+_17281374
|
0.430
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr5_-_146813343
|
0.430
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_-_71307705
|
0.428
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr11_-_94603857
|
0.428
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr5_-_64813459
|
0.425
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr14_+_51525942
|
0.425
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr4_+_183482130
|
0.425
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr22_+_42900168
|
0.424
|
NM_001137605
|
PARVG
|
parvin, gamma
|
|
chr1_-_153109349
|
0.423
|
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr13_-_78075683
|
0.423
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr1_-_68470585
|
0.422
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr18_+_18003401
|
0.420
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr11_+_55319607
|
0.418
|
NM_001004735
|
OR5D14
|
olfactory receptor, family 5, subfamily D, member 14
|
|
chr5_+_76047536
|
0.417
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr4_+_151218862
|
0.415
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr7_-_27171648
|
0.415
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_27206249
|
0.410
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr1_+_84539876
|
0.409
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr6_+_21701942
|
0.408
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr7_+_123036312
|
0.407
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr3_-_186752952
|
0.407
|
NM_139248
|
LIPH
|
lipase, member H
|
|
chr15_-_56829069
|
0.406
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr2_-_159021201
|
0.405
|
NM_001171637
NM_138803
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr4_-_149582948
|
0.405
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr7_-_27171673
|
0.404
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr19_-_6061561
|
0.404
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr3_+_158637273
|
0.404
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr1_+_149299307
|
0.403
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr7_-_27102049
|
0.403
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_-_117300702
|
0.402
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr10_+_22645370
|
0.399
|
|
COMMD3
|
COMM domain containing 3
|
|
chr14_+_51526071
|
0.398
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr7_+_149895390
|
0.397
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr19_+_50086350
|
0.397
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr11_+_6998275
|
0.397
|
NM_176822
|
NLRP14
|
NLR family, pyrin domain containing 14
|
|
chr11_-_123353607
|
0.396
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr1_-_166149867
|
0.395
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr17_-_34607257
|
0.395
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr10_-_127361583
|
0.394
|
NM_001128202
|
C10orf122
|
chromosome 10 open reading frame 122
|
|
chr1_-_148124738
|
0.392
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr14_-_52689680
|
0.391
|
|
DDHD1
|
DDHD domain containing 1
|
|
chr8_-_124622434
|
0.389
|
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr18_-_18002926
|
0.389
|
|
LOC100128893
|
hypothetical LOC100128893
|
|
chr12_+_27511009
|
0.388
|
NM_001145010
|
C12orf70
|
chromosome 12 open reading frame 70
|
|
chr20_+_6696744
|
0.386
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chrX_+_77889861
|
0.385
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr10_+_17311242
|
0.385
|
|
VIM
|
vimentin
|
|
chr19_-_45888259
|
0.382
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr12_-_21702021
|
0.382
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr13_-_45654327
|
0.382
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr5_-_168660293
|
0.381
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr5_-_139402989
|
0.380
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chrX_-_47815944
|
0.379
|
NM_001190255
|
ZNF630
|
zinc finger protein 630
|
|
chr6_+_74462226
|
0.378
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr12_+_105500814
|
0.378
|
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr5_-_137099602
|
0.377
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr12_-_21702046
|
0.376
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr7_-_112513379
|
0.376
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_-_89976261
|
0.376
|
NM_007035
|
KERA
|
keratocan
|
|
chr6_-_85529884
|
0.375
|
|
TBX18
|
T-box 18
|
|
chr12_-_21702036
|
0.374
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr17_+_8154280
|
0.374
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr10_+_11087280
|
0.373
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr12_-_21702000
|
0.371
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr9_-_35101534
|
0.369
|
|
KIAA1539
|
KIAA1539
|
|
chr3_+_174598937
|
0.369
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr7_+_150321771
|
0.369
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr4_-_81213333
|
0.369
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr6_+_74161296
|
0.367
|
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
|
|
chr2_-_2313894
|
0.367
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr3_+_151171755
|
0.366
|
|
LOC646903
|
hypothetical LOC646903
|
|
chr10_+_70881221
|
0.366
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|