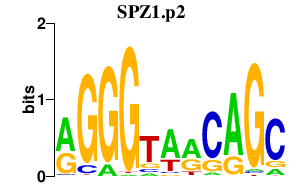
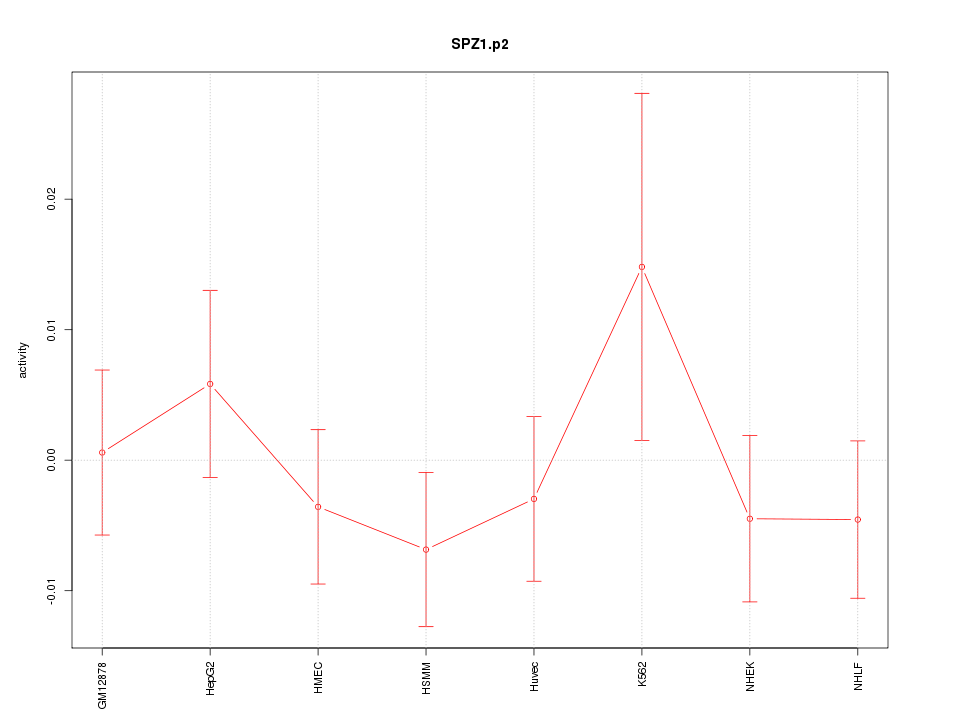
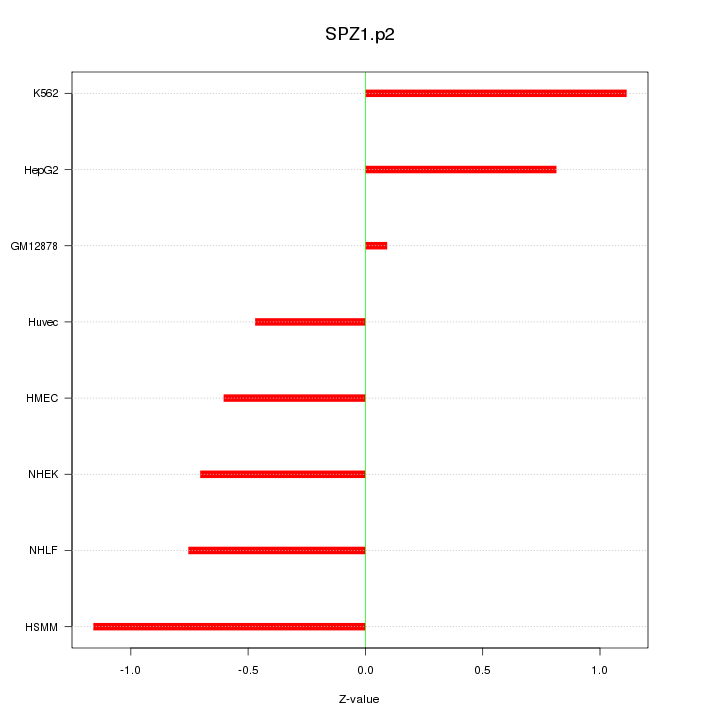
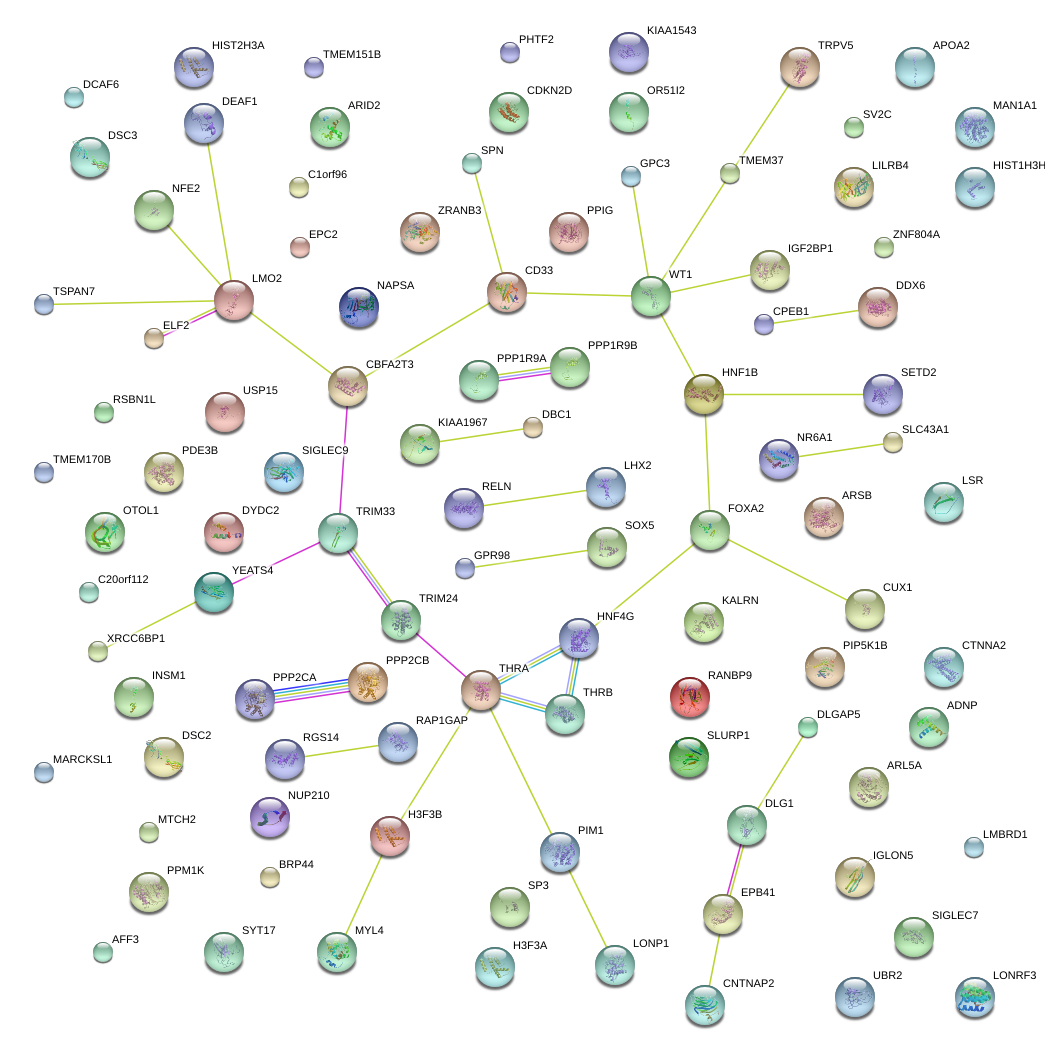
|
chr3_-_13436802
|
1.332
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr20_-_30636535
|
1.255
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr9_+_70509925
|
1.166
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr6_-_119712596
|
1.088
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr18_-_26935883
|
0.943
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_-_32574185
|
0.901
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr3_-_13436725
|
0.899
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr8_+_142471290
|
0.893
|
|
|
|
|
chr2_+_119905812
|
0.826
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr7_-_103417198
|
0.804
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr12_-_52980914
|
0.791
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chrX_-_132947141
|
0.700
|
|
GPC3
|
glypican 3
|
|
chr12_-_52980984
|
0.669
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr12_-_24606616
|
0.669
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr6_+_37245860
|
0.612
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_+_166172679
|
0.609
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chrX_-_132947290
|
0.603
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr1_-_159459999
|
0.554
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr2_-_174537354
|
0.532
|
|
SP3
|
Sp3 transcription factor
|
|
chr2_+_79593679
|
0.528
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr17_+_42641712
|
0.517
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr16_+_29581783
|
0.517
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr7_+_137795719
|
0.509
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr16_-_87535106
|
0.507
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr6_-_13819463
|
0.498
|
|
RANBP9
|
RAN binding protein 9
|
|
chr7_+_77163665
|
0.498
|
NM_198467
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chr5_-_78316891
|
0.480
|
|
ARSB
|
arylsulfatase B
|
|
chr1_-_21868380
|
0.461
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_+_56319948
|
0.459
|
NM_001198558
NM_014441
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9
|
|
chr6_+_42639929
|
0.457
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr2_+_185171480
|
0.454
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr12_+_44409762
|
0.453
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr10_+_82106537
|
0.453
|
NM_032372
|
DYDC2
|
DPY30 domain containing 2
|
|
chr1_+_166172545
|
0.452
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr2_+_149118791
|
0.449
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr11_+_5431213
|
0.447
|
NM_001004754
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2
|
|
chr3_-_198509276
|
0.446
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr22_+_21065134
|
0.443
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chrX_+_117992623
|
0.442
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_+_117992603
|
0.442
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr1_+_29113589
|
0.441
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr16_+_29582069
|
0.433
|
NM_003123
|
SPN
|
sialophorin
|
|
chr16_+_29582065
|
0.429
|
|
SPN
|
sialophorin
|
|
chr16_+_29582480
|
0.423
|
|
SPN
|
sialophorin
|
|
chr2_-_174538197
|
0.421
|
|
SP3
|
Sp3 transcription factor
|
|
chr7_-_142340941
|
0.420
|
NM_019841
|
TRPV5
|
transient receptor potential cation channel, subfamily V, member 5
|
|
chr1_-_181708022
|
0.419
|
|
LOC284649
|
hypothetical LOC284649
|
|
chr2_-_136005238
|
0.412
|
NM_032143
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3
|
|
chr6_-_70563489
|
0.411
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr1_-_227545310
|
0.410
|
NM_145257
|
C1orf96
|
chromosome 1 open reading frame 96
|
|
chr19_-_55560686
|
0.404
|
NM_004851
|
NAPSA
|
napsin A aspartic peptidase
|
|
chr8_-_30789476
|
0.400
|
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr11_-_32413486
|
0.399
|
|
WT1
|
Wilms tumor 1
|
|
chr9_+_125813849
|
0.394
|
|
LHX2
|
LIM homeobox 2
|
|
chr1_+_224317062
|
0.393
|
|
H3F3A
LOC440926
|
H3 histone, family 3A
H3 histone, family 3A pseudogene
|
|
chr12_+_60940404
|
0.391
|
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr2_+_149119029
|
0.384
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr7_+_94374862
|
0.383
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr19_+_56506907
|
0.383
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr1_+_29086206
|
0.383
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr11_-_33847937
|
0.383
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr9_-_126573349
|
0.379
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr17_-_33179118
|
0.378
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_-_183392776
|
0.371
|
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr16_+_19087035
|
0.369
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr7_+_77266017
|
0.368
|
NM_001127358
NM_001127360
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr12_+_68039728
|
0.366
|
NM_006530
|
YEATS4
|
YEATS domain containing 4
|
|
chr11_-_47620590
|
0.361
|
NM_014342
|
MTCH2
|
mitochondrial carrier homolog 2 (C. elegans)
|
|
chr1_-_166172901
|
0.359
|
NM_001143674
|
BRP44
|
brain protein 44
|
|
chr15_-_81113676
|
0.358
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr9_-_121171400
|
0.356
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr20_+_20296744
|
0.352
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr19_+_40431678
|
0.351
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr11_+_14621844
|
0.350
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr4_-_140317686
|
0.346
|
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr1_+_224317036
|
0.346
|
NM_002107
|
H3F3A
|
H3 histone, family 3A
|
|
chr11_+_14621884
|
0.340
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr19_+_7566698
|
0.340
|
NM_001080429
NM_020902
|
KIAA1543
|
KIAA1543
|
|
chr1_-_114855281
|
0.337
|
NM_015906
NM_033020
|
TRIM33
|
tripartite motif containing 33
|
|
chr19_+_56337368
|
0.336
|
NM_014385
NM_016543
|
SIGLEC7
|
sialic acid binding Ig-like lectin 7
|
|
chr2_-_100088691
|
0.336
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr19_-_10540328
|
0.333
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr6_+_44346457
|
0.332
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr2_+_170149348
|
0.330
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr3_+_162697289
|
0.327
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chrX_+_38305634
|
0.324
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr7_+_101247601
|
0.323
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr1_+_224317044
|
0.322
|
|
H3F3A
LOC440926
|
H3 histone, family 3A
H3 histone, family 3A pseudogene
|
|
chr7_+_77266140
|
0.321
|
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr17_+_42641426
|
0.321
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr19_+_56420181
|
0.321
|
|
CD33
|
CD33 molecule
|
|
chr5_+_75414994
|
0.320
|
NM_014979
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
|
chr6_+_11646472
|
0.317
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr7_+_145444385
|
0.316
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr6_-_138231006
|
0.306
|
|
|
|
|
chr15_-_20666684
|
0.305
|
|
LOC283683
|
hypothetical LOC283683
|
|
chr20_-_48980858
|
0.304
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr12_+_56621581
|
0.302
|
|
XRCC6BP1
|
XRCC6 binding protein 1
|
|
chr1_+_224317056
|
0.302
|
|
H3F3A
LOC440926
|
H3 histone, family 3A
H3 histone, family 3A pseudogene
|
|
chr16_+_65701428
|
0.301
|
|
|
|
|
chr8_+_76482661
|
0.300
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr2_-_152393209
|
0.299
|
NM_001037174
NM_012097
NM_177985
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr3_+_125786195
|
0.299
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr19_+_59866237
|
0.299
|
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr17_+_44429748
|
0.296
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr4_-_89424911
|
0.296
|
NM_152542
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr3_-_47180417
|
0.295
|
NM_014159
|
SETD2
|
SET domain containing 2
|
|
chr20_-_22513050
|
0.295
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr3_-_159306214
|
0.295
|
|
|
|
|
chr5_+_176717449
|
0.295
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr5_+_89890366
|
0.294
|
NM_032119
|
GPR98
|
G protein-coupled receptor 98
|
|
chr11_-_57039721
|
0.294
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr12_+_44409743
|
0.293
|
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr11_-_118166797
|
0.292
|
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr3_-_24511456
|
0.291
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr3_-_159932924
|
0.291
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr12_+_49444509
|
0.290
|
|
ATF1
|
activating transcription factor 1
|
|
chr17_-_71352017
|
0.290
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr18_-_50777634
|
0.289
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr17_-_52393364
|
0.288
|
NM_004645
|
COIL
|
coilin
|
|
chr17_-_24302597
|
0.287
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr19_+_12810258
|
0.287
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr1_+_26896519
|
0.287
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr11_-_33848061
|
0.286
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_-_26105477
|
0.286
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr5_+_75734759
|
0.286
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr11_-_33848013
|
0.285
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chrX_-_118711144
|
0.285
|
|
SEPT6
|
septin 6
|
|
chr8_-_74821636
|
0.284
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr2_-_175741096
|
0.283
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr1_+_26609893
|
0.282
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr2_-_152392935
|
0.282
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr19_-_44497815
|
0.282
|
NM_020862
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1
|
|
chr1_+_181708272
|
0.280
|
|
SMG7
|
Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans)
|
|
chr8_-_22841173
|
0.280
|
NM_144962
|
PEBP4
|
phosphatidylethanolamine-binding protein 4
|
|
chr17_-_71352090
|
0.279
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr1_+_29086156
|
0.279
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr12_+_56621711
|
0.278
|
NM_033276
|
XRCC6BP1
|
XRCC6 binding protein 1
|
|
chr6_-_161333363
|
0.277
|
|
|
|
|
chr19_-_10540620
|
0.277
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr6_+_80771040
|
0.277
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr9_-_100510657
|
0.273
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr6_-_70563245
|
0.273
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr1_-_39097904
|
0.272
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr4_-_73653331
|
0.272
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr17_-_73640082
|
0.271
|
NM_007267
|
TMC6
|
transmembrane channel-like 6
|
|
chr4_-_152155827
|
0.270
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chrX_+_70232362
|
0.269
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr1_-_31003181
|
0.268
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr6_+_41856466
|
0.267
|
NM_013397
|
PRICKLE4
|
prickle homolog 4 (Drosophila)
|
|
chr2_-_55774451
|
0.266
|
NM_033109
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1
|
|
chr7_-_100077072
|
0.266
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr5_+_71050745
|
0.265
|
NM_004291
|
CARTPT
|
CART prepropeptide
|
|
chr6_+_31976754
|
0.265
|
NM_001178063
|
C2
|
complement component 2
|
|
chr9_-_126945536
|
0.263
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr6_-_70563564
|
0.262
|
NM_018368
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr6_-_167289659
|
0.261
|
|
|
|
|
chr5_+_137447479
|
0.259
|
NM_058244
|
WNT8A
|
wingless-type MMTV integration site family, member 8A
|
|
chr2_-_152393179
|
0.257
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr18_-_71050268
|
0.256
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr2_+_136005423
|
0.254
|
NM_015361
|
R3HDM1
|
R3H domain containing 1
|
|
chr1_-_32176275
|
0.254
|
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr7_+_138695241
|
0.254
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr6_+_132170888
|
0.253
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr1_+_39229474
|
0.252
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr20_+_60558369
|
0.252
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr2_-_51112743
|
0.252
|
|
NRXN1
|
neurexin 1
|
|
chr5_+_176717553
|
0.251
|
|
|
|
|
chr16_+_51690564
|
0.250
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr1_+_10015736
|
0.249
|
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr19_-_6671585
|
0.249
|
NM_000064
|
C3
|
complement component 3
|
|
chr7_+_30291136
|
0.248
|
|
ZNRF2
|
zinc and ring finger 2
|
|
chr16_-_51138306
|
0.248
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr15_-_35178888
|
0.248
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr14_-_37133927
|
0.248
|
|
FOXA1
|
forkhead box A1
|
|
chr3_+_157071173
|
0.247
|
|
GMPS
|
guanine monphosphate synthetase
|
|
chr18_-_802268
|
0.247
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr2_-_70848621
|
0.247
|
NM_001185055
NM_001617
NM_017482
NM_017488
|
ADD2
|
adducin 2 (beta)
|
|
chr22_+_20252020
|
0.246
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr6_-_114398946
|
0.246
|
|
HDAC2
|
histone deacetylase 2
|
|
chr7_-_122961808
|
0.246
|
NM_178827
|
IQUB
|
IQ motif and ubiquitin domain containing
|
|
chr7_+_100647680
|
0.246
|
NM_006349
|
ZNHIT1
|
zinc finger, HIT-type containing 1
|
|
chr2_-_192767844
|
0.246
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr3_+_12501224
|
0.245
|
|
TSEN2
|
tRNA splicing endonuclease 2 homolog (S. cerevisiae)
|
|
chr18_+_30812205
|
0.244
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr22_+_20252004
|
0.242
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chrX_+_48529905
|
0.242
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr1_-_31003260
|
0.241
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr7_+_21434097
|
0.241
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr10_+_49562866
|
0.241
|
|
WDFY4
|
WDFY family member 4
|
|
chr12_-_42486314
|
0.241
|
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr1_-_180627023
|
0.240
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chrX_-_13866366
|
0.240
|
|
|
|
|
chr3_-_95175282
|
0.238
|
|
PROS1
|
protein S (alpha)
|
|
chr1_-_180627934
|
0.238
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr11_+_117812282
|
0.238
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr10_+_35456095
|
0.238
|
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_75962959
|
0.238
|
|
ACADM
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain
|
|
chr1_-_78217360
|
0.235
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr1_+_170768635
|
0.234
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|