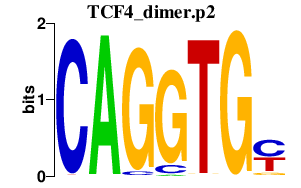
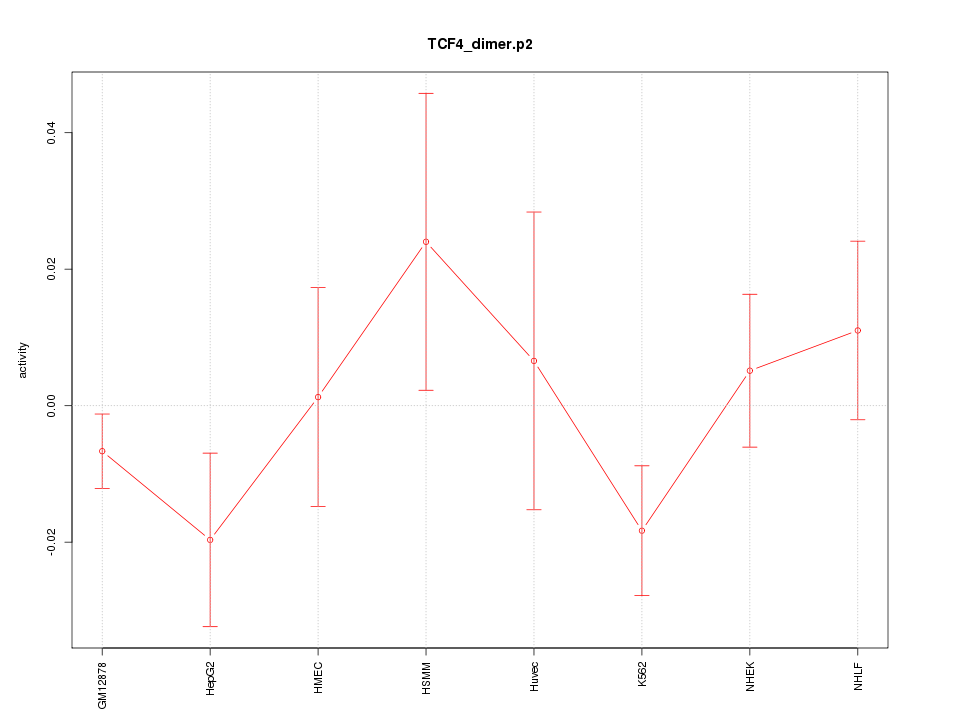
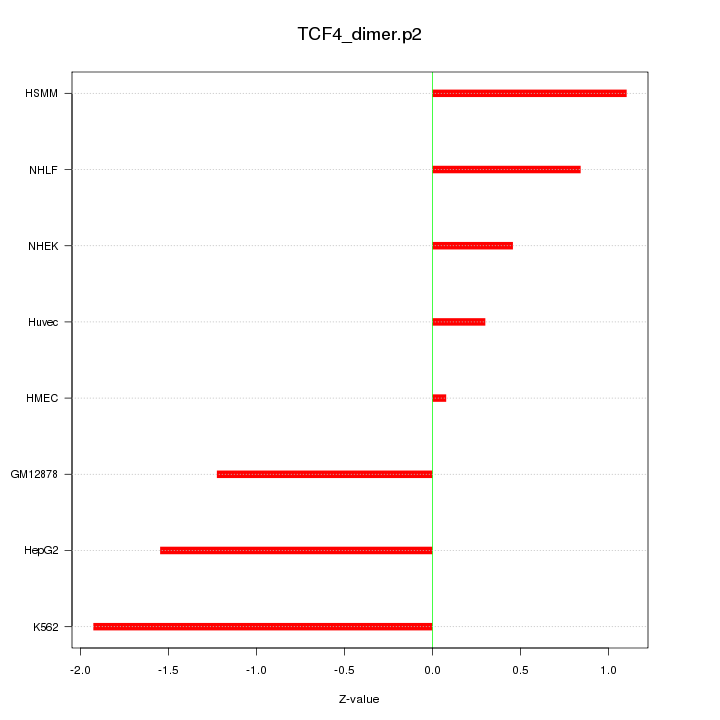
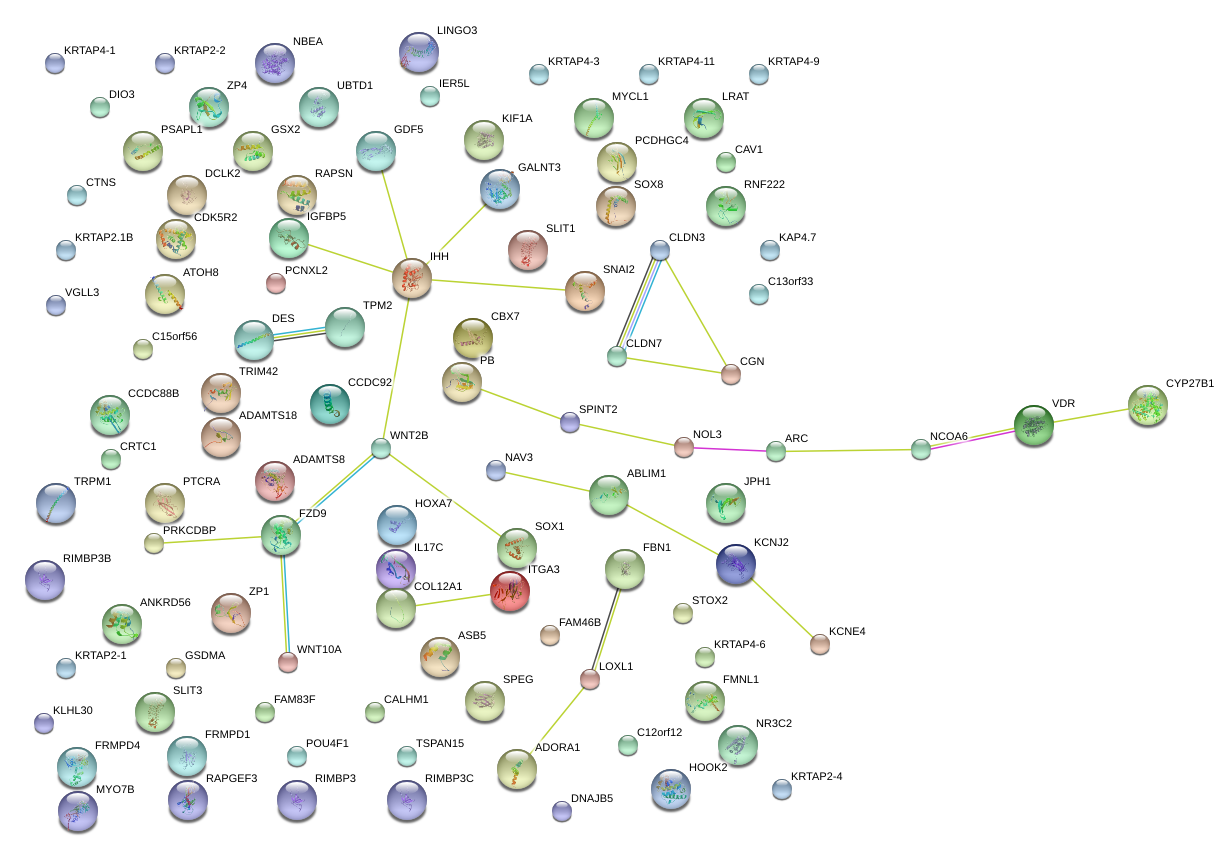
|
chr19_+_43446937
|
4.416
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_43447078
|
4.328
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_18655487
|
4.220
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr19_+_43447262
|
3.937
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr9_-_130980359
|
3.540
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr1_+_149750485
|
3.155
|
NM_020770
|
CGN
|
cingulin
|
|
chr2_+_85834111
|
3.116
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr1_+_201364050
|
3.052
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr8_-_49996353
|
3.008
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr1_-_27211913
|
2.987
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr3_-_87122936
|
2.819
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr14_+_101097440
|
2.755
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr19_+_18655483
|
2.744
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr2_-_241408296
|
2.676
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr5_+_140844810
|
2.596
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr11_+_60391590
|
2.587
|
NM_207341
|
ZP1
|
zona pellucida glycoprotein 1 (sperm receptor)
|
|
chr1_-_231498081
|
2.498
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr9_-_130980302
|
2.483
|
|
IER5L
|
immediate early response 5-like
|
|
chr4_+_54660954
|
2.473
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr22_+_20068039
|
2.439
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr17_+_45488356
|
2.382
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr11_-_47427263
|
2.367
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr22_+_38720881
|
2.352
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr6_+_42991704
|
2.350
|
NM_138296
|
PTCRA
|
pre T-cell antigen receptor alpha
|
|
chr13_+_30378311
|
2.318
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr10_+_99248605
|
2.292
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr17_-_36577949
|
2.279
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr17_+_45488330
|
2.259
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr8_-_75396016
|
2.242
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr17_-_36594652
|
2.198
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr22_-_37858871
|
2.174
|
|
CBX7
|
chromobox homolog 7
|
|
chr10_-_105208634
|
2.146
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr1_-_27212035
|
2.112
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr2_+_128009782
|
2.070
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr15_-_38332401
|
2.065
|
NM_001039905
|
C15orf56
|
chromosome 15 open reading frame 56
|
|
chr4_-_177427252
|
2.003
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr15_-_46725087
|
1.993
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr17_-_36528113
|
1.977
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr11_-_129803712
|
1.965
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr19_-_2259155
|
1.941
|
NM_001101391
|
LINGO3
|
leucine rich repeat and Ig domain containing 3
|
|
chr17_+_36493927
|
1.938
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr11_-_129803515
|
1.900
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr11_-_6298284
|
1.856
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_-_40140126
|
1.815
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr2_-_219633481
|
1.812
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr2_+_219991342
|
1.798
|
NM_001927
|
DES
|
desmin
|
|
chr16_+_971808
|
1.790
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr16_+_972064
|
1.786
|
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chrX_+_12066505
|
1.782
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr3_-_87122958
|
1.782
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr7_+_115953582
|
1.776
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr10_-_116517945
|
1.764
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_+_238712101
|
1.761
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr13_-_78075683
|
1.750
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr4_-_149582948
|
1.747
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr2_+_223625105
|
1.728
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr6_-_75972286
|
1.728
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr10_+_70881221
|
1.716
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr7_-_72822471
|
1.703
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr2_-_166358951
|
1.696
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr17_+_40655060
|
1.686
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr4_+_185063421
|
1.676
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr17_+_3486510
|
1.664
|
NM_001031681
NM_004937
|
CTNS
|
cystinosis, nephropathic
|
|
chr2_+_220007943
|
1.664
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr12_+_76749199
|
1.664
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr2_+_219453487
|
1.659
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr15_+_72005839
|
1.641
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_35372751
|
1.638
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr12_-_123023312
|
1.633
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr6_-_75972473
|
1.631
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr17_-_8241868
|
1.628
|
NM_001146684
|
RNF222
|
ring finger protein 222
|
|
chr1_+_112852892
|
1.622
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr4_-_7487514
|
1.621
|
NM_001085382
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene)
|
|
chr16_+_87232501
|
1.614
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr2_+_219532590
|
1.612
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr15_+_72005847
|
1.604
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_72006015
|
1.595
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr5_-_168660293
|
1.587
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr17_-_7106987
|
1.577
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr20_-_33489383
|
1.575
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr7_-_27162688
|
1.575
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr9_+_37640996
|
1.573
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr3_+_141879555
|
1.548
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr2_-_217267742
|
1.539
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr7_+_72486044
|
1.532
|
NM_003508
|
FZD9
|
frizzled homolog 9 (Drosophila)
|
|
chr17_+_65677255
|
1.524
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr4_+_151218862
|
1.522
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr13_+_111769913
|
1.521
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr19_-_12747263
|
1.504
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr4_+_155884564
|
1.501
|
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr12_-_46438447
|
1.486
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr8_-_143692814
|
1.482
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr12_-_46585032
|
1.455
|
NM_000376
NM_001017535
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr4_-_78038025
|
1.444
|
NM_001029870
|
ANKRD56
|
ankyrin repeat domain 56
|
|
chr16_-_76026431
|
1.437
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr11_+_63864263
|
1.434
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr15_-_29181198
|
1.431
|
NM_002420
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1
|
|
chr9_-_35679902
|
1.411
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr1_-_153107084
|
1.409
|
|
|
|
|
chr13_+_34414390
|
1.402
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr12_-_89873081
|
1.385
|
NM_152638
|
C12orf12
|
chromosome 12 open reading frame 12
|
|
chr9_+_34980266
|
1.381
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr17_-_36456949
|
1.374
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr17_-_70480345
|
1.360
|
|
|
|
|
chr2_-_175337386
|
1.351
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr21_+_46355797
|
1.349
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_-_47679176
|
1.343
|
NM_015086
|
DDN
|
dendrin
|
|
chr2_-_37752731
|
1.338
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr3_-_123223658
|
1.338
|
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr2_+_205118961
|
1.334
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr10_+_115988999
|
1.327
|
NM_198496
|
VWA2
|
von Willebrand factor A domain containing 2
|
|
chr1_-_42157077
|
1.320
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr9_-_135379888
|
1.312
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr3_-_39209075
|
1.307
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr15_-_23659329
|
1.299
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr8_+_1909437
|
1.298
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr14_+_104402626
|
1.292
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr11_+_70915960
|
1.291
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr11_+_6237541
|
1.283
|
NM_176875
|
CCKBR
|
cholecystokinin B receptor
|
|
chr15_-_73818013
|
1.282
|
|
|
|
|
chr8_-_29262240
|
1.279
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr11_-_118055564
|
1.275
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chrX_-_128616594
|
1.273
|
NM_017413
|
APLN
|
apelin
|
|
chr14_+_104852225
|
1.272
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr1_-_42156957
|
1.267
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr20_-_29896981
|
1.264
|
NM_004118
|
FOXS1
|
forkhead box S1
|
|
chr11_-_67163600
|
1.258
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr3_+_128874458
|
1.257
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr16_+_1143738
|
1.256
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr16_+_67236699
|
1.254
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr9_-_109291130
|
1.246
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr10_-_90957028
|
1.243
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr8_-_139995417
|
1.242
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr1_+_177978920
|
1.242
|
NM_173509
|
FAM163A
|
family with sequence similarity 163, member A
|
|
chr3_-_64186151
|
1.233
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr17_+_1905104
|
1.231
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr10_+_99463446
|
1.224
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr1_-_2451543
|
1.223
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr19_-_61874934
|
1.218
|
|
ZNF835
|
zinc finger protein 835
|
|
chr16_+_56230707
|
1.212
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr5_-_176832476
|
1.211
|
NM_080881
|
DBN1
|
drebrin 1
|
|
chr6_-_39398293
|
1.199
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr16_+_67236730
|
1.197
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chrX_+_71047344
|
1.194
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr17_+_58058774
|
1.190
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr1_-_3517918
|
1.190
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr5_-_146238336
|
1.188
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr16_+_29697090
|
1.184
|
|
ZG16
|
zymogen granule protein 16 homolog (rat)
|
|
chr10_+_103338030
|
1.174
|
NM_015448
|
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse)
|
|
chr2_+_120820140
|
1.163
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr9_-_139060285
|
1.162
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_-_39877883
|
1.156
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr3_+_85090721
|
1.155
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr22_-_36212160
|
1.149
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr19_+_18655391
|
1.146
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr3_+_42702014
|
1.145
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr10_+_99463468
|
1.144
|
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr2_-_189752575
|
1.141
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr19_+_10597167
|
1.133
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr10_-_99521701
|
1.132
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr8_-_144862266
|
1.131
|
NM_001162914
|
LOC100130274
|
coiled-coil domain containing 121-like
|
|
chr11_-_65070625
|
1.131
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr21_-_38210531
|
1.125
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr16_-_63713466
|
1.124
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr19_+_45664889
|
1.124
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr22_+_39096595
|
1.118
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr17_+_71586857
|
1.114
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr17_+_24944605
|
1.104
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr12_-_68379331
|
1.102
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr1_+_199975271
|
1.101
|
|
NAV1
|
neuron navigator 1
|
|
chr6_-_33822659
|
1.100
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr8_+_144888275
|
1.099
|
|
LOC100128338
|
hypothetical LOC100128338
|
|
chr9_+_101623957
|
1.098
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_-_158182006
|
1.096
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr18_-_55091596
|
1.093
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr12_+_6179736
|
1.093
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr9_-_35679799
|
1.089
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr2_-_216944911
|
1.087
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr19_-_18493847
|
1.080
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr19_-_18493886
|
1.080
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr11_+_122936246
|
1.080
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr20_+_1194959
|
1.078
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr11_+_36354110
|
1.077
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr19_+_43502380
|
1.076
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr11_+_120478584
|
1.069
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr5_+_74197473
|
1.067
|
|
LOC441086
|
hypothetical LOC441086
|
|
chr9_-_96441654
|
1.066
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr10_-_98935369
|
1.066
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr5_+_94981735
|
1.065
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr19_-_7845325
|
1.062
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr1_-_109458001
|
1.060
|
NM_001122961
|
C1orf194
|
chromosome 1 open reading frame 194
|
|
chr11_-_67032125
|
1.058
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr1_-_39871226
|
1.054
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr16_+_21603250
|
1.053
|
NM_001161683
|
OTOA
|
otoancorin
|
|
chr7_+_75947762
|
1.049
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr16_-_73012740
|
1.046
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr2_-_192767494
|
1.044
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr1_-_109853808
|
1.044
|
NM_020703
|
AMIGO1
|
adhesion molecule with Ig-like domain 1
|
|
chr9_-_139042526
|
1.044
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_-_84237316
|
1.043
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|