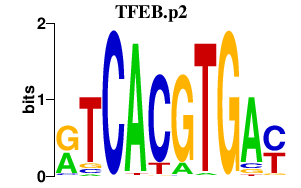
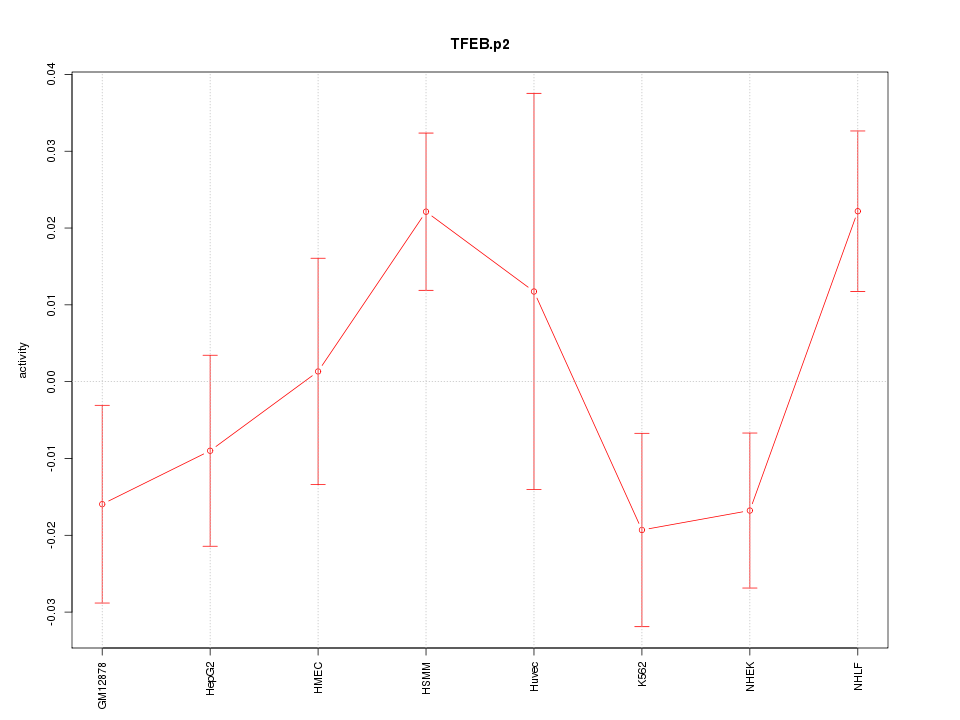
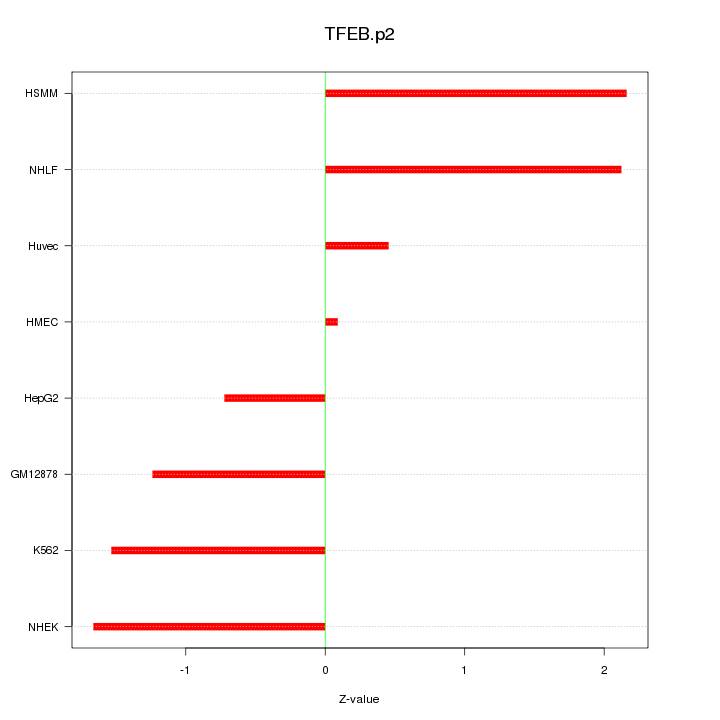
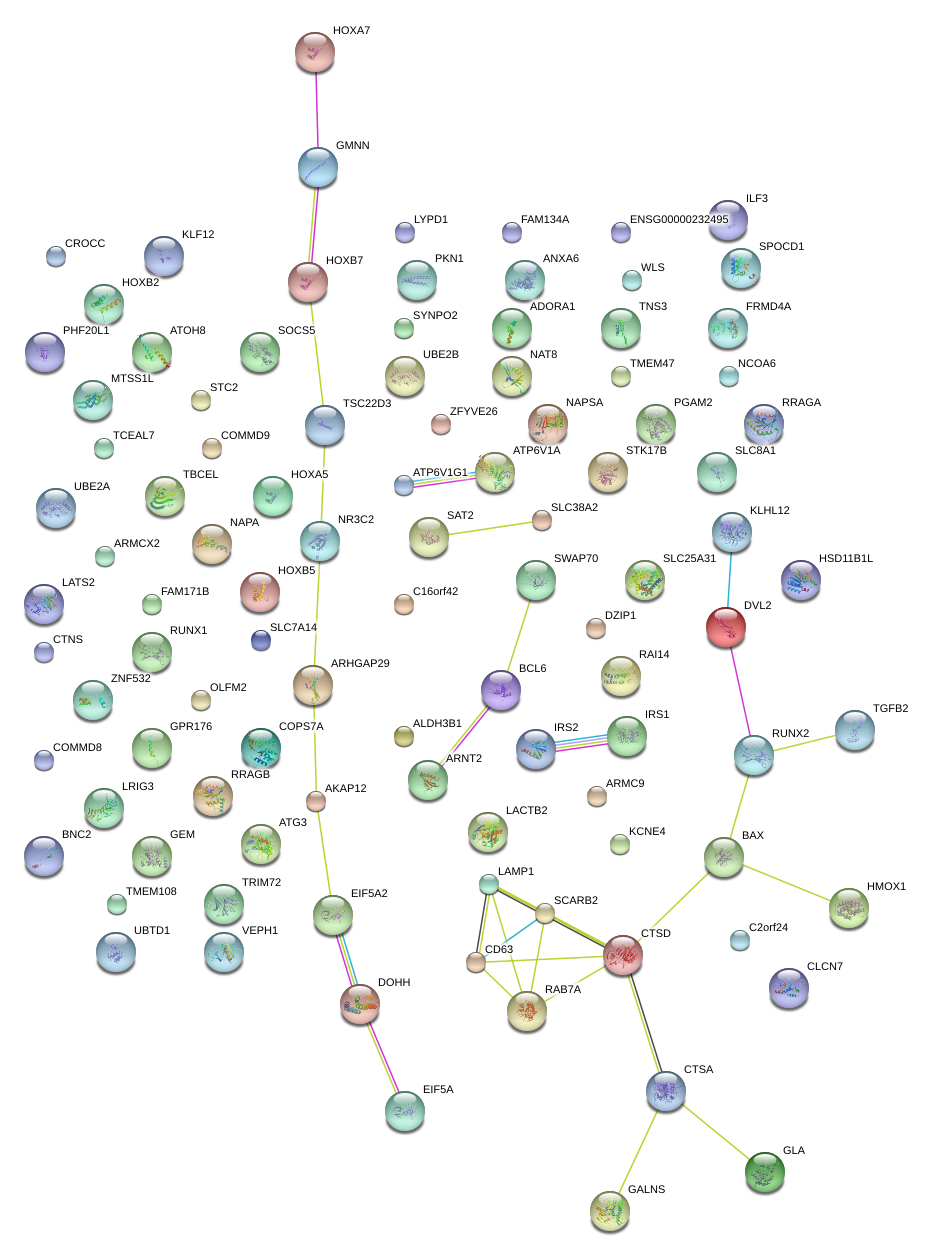
|
chr13_+_112999538
|
2.703
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr17_-_44043361
|
2.410
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr13_+_112999590
|
2.326
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_112999462
|
2.306
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chrX_-_100801472
|
2.027
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr7_-_27162688
|
1.815
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr3_-_113763454
|
1.679
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr2_+_85834111
|
1.613
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr1_-_197173159
|
1.412
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chrX_+_55760774
|
1.397
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr17_-_7078399
|
1.378
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr1_-_68470585
|
1.372
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr5_+_34692124
|
1.367
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr4_+_120167721
|
1.311
|
|
SYNPO2
|
synaptopodin 2
|
|
chr22_+_34107087
|
1.287
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr8_-_95343716
|
1.257
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_-_95343696
|
1.248
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_-_40510923
|
1.246
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr19_-_52710129
|
1.242
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr9_-_16860719
|
1.204
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr8_-_71743908
|
1.190
|
|
LACTB2
|
lactamase, beta 2
|
|
chr15_-_38000379
|
1.188
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chrX_-_34585287
|
1.184
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr13_-_95094920
|
1.184
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr5_+_34692239
|
1.175
|
|
RAI14
|
retinoic acid induced 14
|
|
chr17_-_7078446
|
1.164
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr22_+_34107051
|
1.157
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr4_+_128870968
|
1.155
|
NM_031291
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chr11_+_120399966
|
1.144
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr9_-_16860664
|
1.133
|
|
BNC2
|
basonuclin 2
|
|
chr17_+_3486753
|
1.130
|
|
CTNS
|
cystinosis, nephropathic
|
|
chr1_-_94475840
|
1.103
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr7_-_47588652
|
1.090
|
|
TNS3
|
tensin 3
|
|
chr1_-_32054164
|
1.083
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr2_+_223625105
|
1.061
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr3_-_113763449
|
1.055
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr4_+_120167770
|
1.051
|
|
SYNPO2
|
synaptopodin 2
|
|
chr5_+_34692352
|
1.048
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr16_-_1341766
|
1.032
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr17_-_44042924
|
1.028
|
|
HOXB7
|
homeobox B7
|
|
chr10_-_14090459
|
1.023
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_-_47588160
|
1.006
|
|
TNS3
|
tensin 3
|
|
chr19_+_10625896
|
1.002
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr4_-_47160476
|
0.996
|
|
COMMD8
|
COMM domain containing 8
|
|
chr16_-_87450734
|
0.974
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr13_-_73605914
|
0.959
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr2_+_219751145
|
0.950
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr16_+_31132842
|
0.939
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr1_+_216585298
|
0.935
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr19_+_5632139
|
0.927
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr2_-_227372574
|
0.926
|
|
IRS1
|
insulin receptor substrate 1
|
|
chrX_-_100549444
|
0.922
|
|
GLA
|
galactosidase, alpha
|
|
chr8_-_71743973
|
0.922
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr8_+_133856738
|
0.918
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr19_-_10625449
|
0.915
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr2_+_187266899
|
0.895
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr17_-_7471832
|
0.887
|
NM_133491
|
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2
|
|
chr3_+_108442205
|
0.878
|
|
LOC344595
|
hypothetical LOC344595
|
|
chr3_-_188946168
|
0.877
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr16_-_69277355
|
0.871
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr2_+_46779553
|
0.869
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr20_+_43953372
|
0.860
|
|
CTSA
|
cathepsin A
|
|
chr2_-_133144094
|
0.860
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr19_-_3451620
|
0.851
|
NM_031304
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase
|
|
chr11_+_9642200
|
0.833
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr12_-_57600336
|
0.827
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr17_-_7078304
|
0.826
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr12_-_57600564
|
0.825
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr17_-_44026042
|
0.819
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr3_+_129927714
|
0.810
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr1_+_201364050
|
0.805
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr17_-_43977273
|
0.803
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr11_+_9642226
|
0.799
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr2_-_196744546
|
0.799
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chrX_+_102471769
|
0.797
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr3_+_129927668
|
0.789
|
NM_004637
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr11_-_36267510
|
0.785
|
|
COMMD9
|
COMM domain containing 9
|
|
chr11_+_9642182
|
0.783
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr1_-_201162991
|
0.780
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr3_+_129927723
|
0.777
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr2_+_219751192
|
0.773
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr3_+_129927716
|
0.768
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr10_+_99248605
|
0.766
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr14_-_67353046
|
0.763
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr3_-_158700036
|
0.762
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr4_-_149582948
|
0.760
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr5_-_150517499
|
0.759
|
|
ANXA6
|
annexin A6
|
|
chr12_-_54409059
|
0.756
|
|
CD63
|
CD63 molecule
|
|
chr4_-_77354011
|
0.755
|
NM_005506
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr3_+_114948550
|
0.753
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr3_-_172109119
|
0.750
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr18_+_54682261
|
0.750
|
|
ZNF532
|
zinc finger protein 532
|
|
chr13_-_109236897
|
0.743
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr21_-_35343448
|
0.741
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_+_67532592
|
0.739
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr7_-_44071640
|
0.725
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr7_-_27149750
|
0.725
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr2_+_231771622
|
0.719
|
|
ARMC9
|
armadillo repeat containing 9
|
|
chr13_-_20533703
|
0.715
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr13_-_20533629
|
0.713
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_+_231771584
|
0.713
|
NM_025139
|
ARMC9
|
armadillo repeat containing 9
|
|
chr2_+_219751230
|
0.713
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr11_+_9642229
|
0.711
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr6_+_151602787
|
0.711
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_-_1741680
|
0.707
|
|
CTSD
|
cathepsin D
|
|
chr21_-_35343456
|
0.706
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_35343500
|
0.705
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_-_219750975
|
0.704
|
|
C2orf24
|
chromosome 2 open reading frame 24
|
|
chr5_-_172687798
|
0.704
|
|
STC2
|
stanniocalcin 2
|
|
chr1_+_17121012
|
0.700
|
NM_014675
|
CROCC
|
ciliary rootlet coiled-coil, rootletin
|
|
chr19_+_54149843
|
0.699
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr3_-_188945921
|
0.697
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_+_14405100
|
0.696
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr5_+_133734768
|
0.694
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr3_+_134239860
|
0.693
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chrX_-_106846924
|
0.690
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_+_14405216
|
0.690
|
|
PKN1
|
protein kinase N1
|
|
chr9_+_116389918
|
0.690
|
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr7_+_27102197
|
0.689
|
|
|
|
|
chr7_+_27102508
|
0.687
|
|
|
|
|
chr12_+_6703686
|
0.686
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr5_+_148766632
|
0.686
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr6_+_151603201
|
0.685
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr16_-_1465021
|
0.685
|
|
CLCN7
|
chloride channel 7
|
|
chr3_-_171786468
|
0.684
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr19_-_9908014
|
0.679
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr7_-_47588266
|
0.679
|
|
TNS3
|
tensin 3
|
|
chr13_+_48582389
|
0.675
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_-_44022595
|
0.674
|
|
HOXB3
|
homeobox B3
|
|
chr6_+_151603264
|
0.671
|
|
|
|
|
chr12_+_54396099
|
0.669
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr2_+_238060602
|
0.665
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr5_+_176663385
|
0.665
|
|
PRELID1
|
PRELI domain containing 1
|
|
chr10_+_70553993
|
0.664
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr17_-_33043514
|
0.660
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr1_-_202595666
|
0.660
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr3_-_49370755
|
0.659
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr11_-_47427263
|
0.655
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr19_+_54708389
|
0.653
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr1_-_203557379
|
0.652
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr11_-_1741663
|
0.652
|
|
CTSD
|
cathepsin D
|
|
chr7_+_17304707
|
0.652
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chrX_+_10084984
|
0.649
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr14_+_102458745
|
0.649
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr16_-_57276108
|
0.648
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr11_+_101485946
|
0.647
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr9_-_89779180
|
0.645
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr16_-_71549149
|
0.644
|
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_-_54918369
|
0.640
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr7_+_27191534
|
0.639
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr5_+_72957736
|
0.637
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr1_-_151775221
|
0.632
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr10_+_102746819
|
0.631
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr9_+_116389814
|
0.628
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr18_+_75967902
|
0.625
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr3_-_49370682
|
0.618
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr9_-_135379888
|
0.617
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr2_+_238712101
|
0.615
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr2_+_231771623
|
0.612
|
|
ARMC9
|
armadillo repeat containing 9
|
|
chr11_+_119701047
|
0.608
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chrX_+_101862297
|
0.608
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chrX_+_100692071
|
0.606
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr22_+_34025872
|
0.605
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr2_+_148495043
|
0.604
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr5_-_150517454
|
0.603
|
|
ANXA6
|
annexin A6
|
|
chr8_+_1759502
|
0.602
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr3_+_133619193
|
0.599
|
NM_015268
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13
|
|
chr1_-_152797820
|
0.595
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr12_-_108918570
|
0.594
|
NM_001135213
NM_001135214
NM_014776
NM_057169
NM_057170
NM_139201
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr18_-_41801237
|
0.594
|
NM_020964
|
EPG5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans)
|
|
chr17_-_74487527
|
0.594
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr13_-_35818603
|
0.589
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr17_+_75689955
|
0.588
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr17_-_44010543
|
0.587
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr4_-_164473107
|
0.585
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr12_-_54409067
|
0.585
|
|
CD63
|
CD63 molecule
|
|
chr1_-_94475504
|
0.581
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr12_-_108918440
|
0.581
|
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr13_-_49057707
|
0.581
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr9_-_139128979
|
0.579
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr17_+_18159294
|
0.576
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr14_+_19993181
|
0.575
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr16_-_1465007
|
0.575
|
|
CLCN7
|
chloride channel 7
|
|
chr14_-_91642703
|
0.574
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr11_+_66915997
|
0.573
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr8_-_133140808
|
0.572
|
NM_001080399
|
OC90
|
otoconin 90
|
|
chr4_-_95482994
|
0.571
|
NM_014485
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr12_-_47604930
|
0.570
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr1_+_181871830
|
0.565
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_-_169392771
|
0.564
|
|
|
|
|
chr1_-_153477673
|
0.563
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr1_+_44213229
|
0.562
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr15_+_46200460
|
0.561
|
NM_205850
|
SLC24A5
|
solute carrier family 24, member 5
|
|
chr19_-_5291700
|
0.558
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr12_+_52806103
|
0.555
|
|
LOC400043
|
hypothetical LOC400043
|
|
chr3_-_172109075
|
0.555
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_+_185515013
|
0.550
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_-_153477511
|
0.546
|
|
GBA
|
glucosidase, beta, acid
|
|
chr3_+_160002605
|
0.543
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_152797760
|
0.543
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|