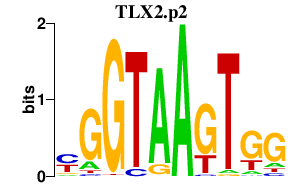
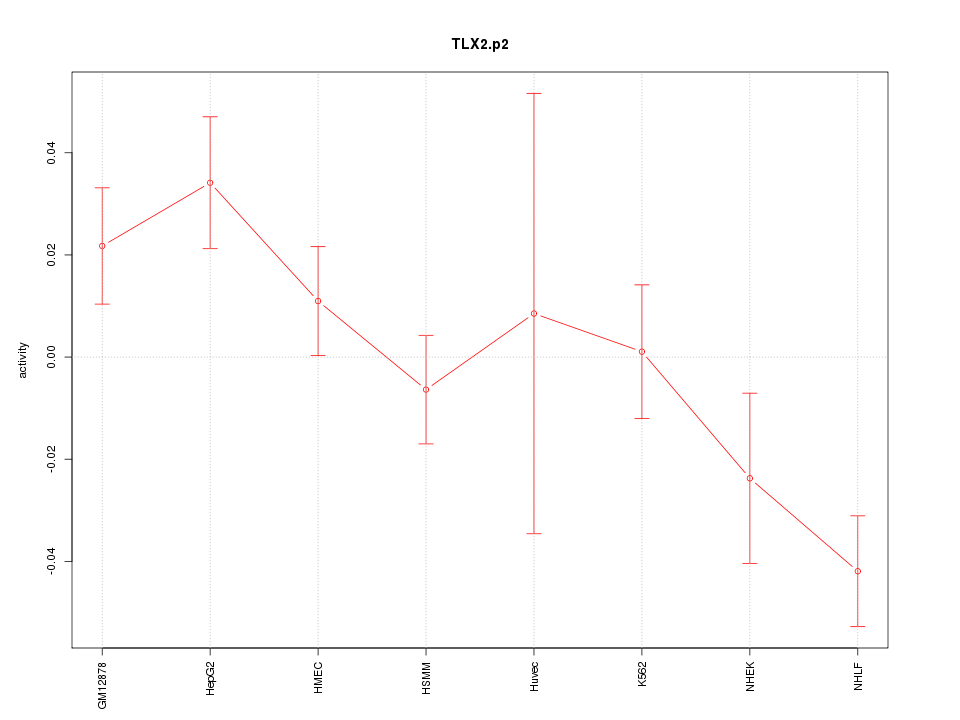
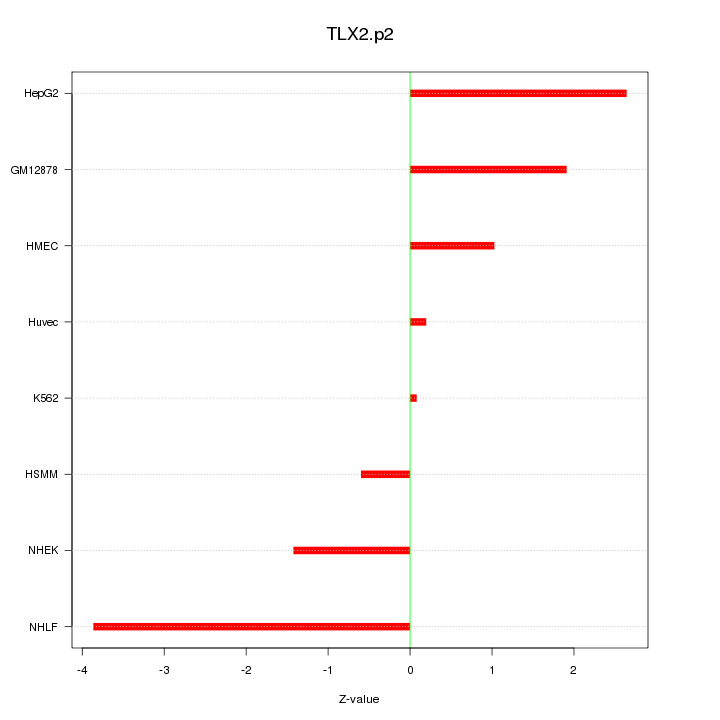
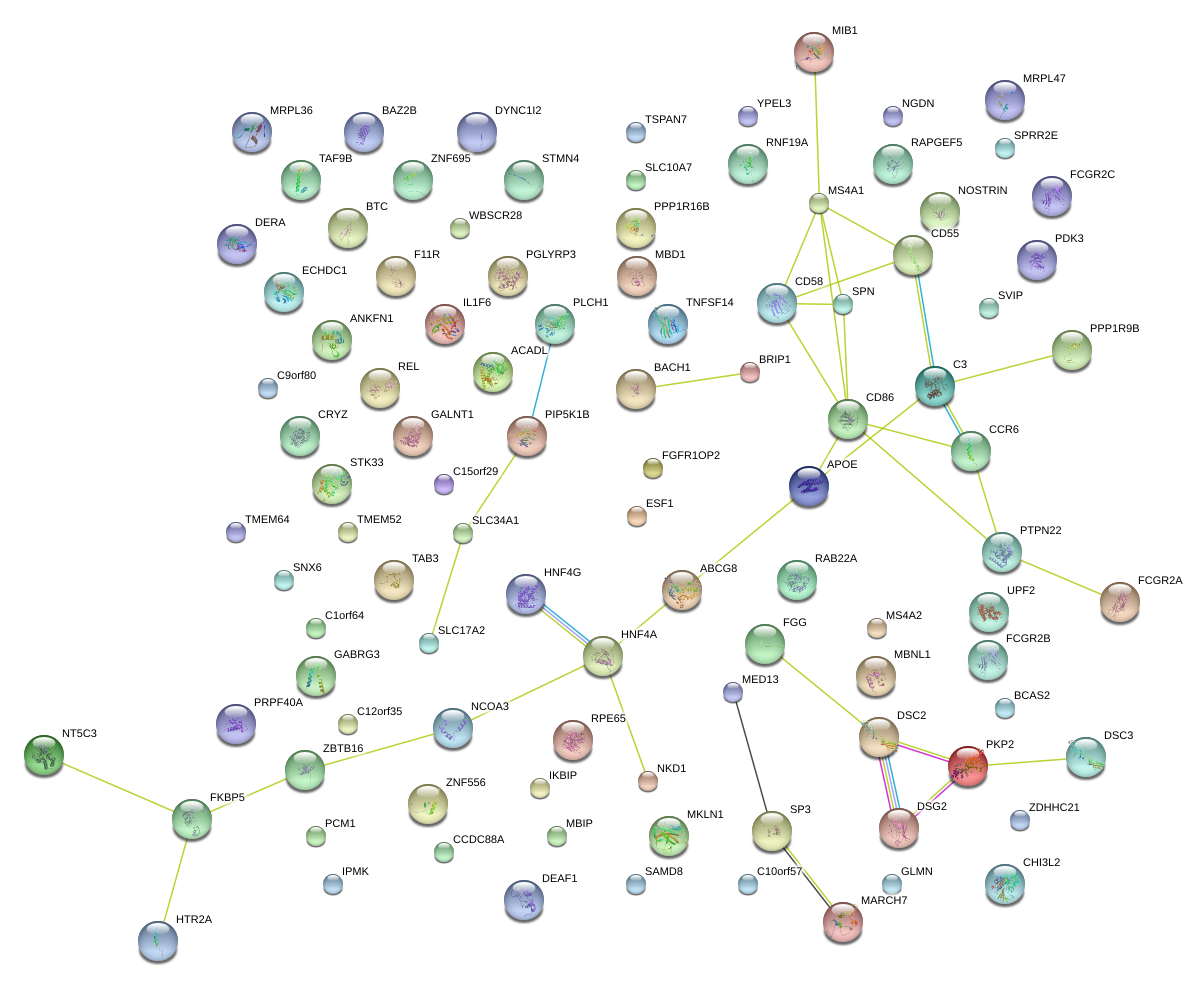
|
chr11_-_8571988
|
1.209
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr4_-_75938710
|
1.148
|
NM_001729
|
BTC
|
betacellulin
|
|
chr13_-_46368968
|
1.117
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr9_+_70509925
|
1.111
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr1_-_245237977
|
1.019
|
NM_020394
|
ZNF695
|
zinc finger protein 695
|
|
chrX_+_38305634
|
0.991
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr7_-_22363036
|
0.940
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr6_+_167445284
|
0.917
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr1_-_68688219
|
0.910
|
NM_000329
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa
|
|
chr6_-_26038817
|
0.887
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr1_-_245237925
|
0.883
|
|
ZNF695
|
zinc finger protein 695
|
|
chr3_+_123279439
|
0.864
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr12_-_97562532
|
0.864
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr11_-_22807842
|
0.860
|
NM_148893
|
SVIP
|
small VCP/p97-interacting protein
|
|
chr2_+_43919606
|
0.807
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr14_-_35859545
|
0.799
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr20_+_42463350
|
0.796
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_+_42463387
|
0.771
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_+_56318415
|
0.754
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr18_+_27332024
|
0.742
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr1_+_16203317
|
0.734
|
NM_178840
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr10_+_82163568
|
0.727
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr20_+_42463419
|
0.722
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr10_-_59697636
|
0.720
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr18_-_26935883
|
0.719
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_-_74971276
|
0.717
|
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr10_-_12125028
|
0.716
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr16_+_49139694
|
0.700
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr2_-_160277083
|
0.696
|
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr1_+_159817752
|
0.694
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr11_-_8572321
|
0.690
|
|
STK33
|
serine/threonine kinase 33
|
|
chrX_-_77281764
|
0.681
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr2_+_172252194
|
0.671
|
NM_001378
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr3_+_153469421
|
0.671
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr12_-_32940931
|
0.664
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr6_-_127706204
|
0.660
|
|
ECHDC1
|
enoyl CoA hydratase domain containing 1
|
|
chr6_-_35804337
|
0.646
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr2_-_210798232
|
0.646
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr11_-_8572277
|
0.644
|
|
STK33
|
serine/threonine kinase 33
|
|
chr10_-_12124778
|
0.643
|
NM_015542
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chrX_-_30817329
|
0.641
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr7_+_72913424
|
0.640
|
NM_182504
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr2_+_169367345
|
0.637
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr7_-_112545840
|
0.637
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr11_+_59979857
|
0.636
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr1_-_114925734
|
0.632
|
NM_005872
|
BCAS2
|
breast carcinoma amplified sequence 2
|
|
chr2_+_60962279
|
0.628
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr20_-_13713509
|
0.627
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr17_+_51585834
|
0.625
|
NM_153228
|
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1
|
|
chr17_-_57497380
|
0.623
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr4_-_155753249
|
0.622
|
|
FGG
|
fibrinogen gamma chain
|
|
chr8_-_91726915
|
0.622
|
|
TMEM64
|
transmembrane protein 64
|
|
chr20_+_47328585
|
0.614
|
|
NCRNA00275
|
non-protein coding RNA 275
|
|
chr17_+_55269974
|
0.614
|
|
|
|
|
chr7_-_112545847
|
0.609
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr1_-_114215768
|
0.604
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr7_-_33047042
|
0.603
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr3_-_180805104
|
0.592
|
NM_020409
NM_177988
|
MRPL47
|
mitochondrial ribosomal protein L47
|
|
chr2_-_174537021
|
0.589
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr11_+_113435640
|
0.585
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_-_92537087
|
0.584
|
NM_053274
|
GLMN
|
glomulin, FKBP associated protein
|
|
chr20_+_42417502
|
0.580
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr15_+_24799174
|
0.577
|
NM_033223
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3
|
|
chr18_+_31488530
|
0.574
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr8_+_76614757
|
0.574
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr12_+_15955549
|
0.571
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr19_-_6671585
|
0.566
|
NM_000064
|
C3
|
complement component 3
|
|
chr1_-_151549790
|
0.563
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr14_+_23008742
|
0.561
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr1_-_116915163
|
0.555
|
NM_001144822
NM_001779
|
CD58
|
CD58 molecule
|
|
chr1_+_111573855
|
0.553
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chr4_-_147662318
|
0.551
|
NM_001029998
NM_032128
|
SLC10A7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7
|
|
chr2_+_160277219
|
0.551
|
NM_022826
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr12_+_26982571
|
0.550
|
NM_001171887
NM_001171888
NM_015633
|
FGFR1OP2
|
FGFR1 oncogene partner 2
|
|
chr1_-_159257486
|
0.547
|
|
F11R
|
F11 receptor
|
|
chr1_-_1840564
|
0.544
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr9_-_114520118
|
0.543
|
|
C9orf80
|
chromosome 9 open reading frame 80
|
|
chr14_-_34168911
|
0.541
|
|
SNX6
|
sorting nexin 6
|
|
chr16_+_29581783
|
0.541
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr18_+_58006154
|
0.541
|
|
|
|
|
chr15_-_32289561
|
0.540
|
NM_024713
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr20_+_45564007
|
0.539
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr8_+_17874128
|
0.539
|
|
PCM1
|
pericentriolar material 1
|
|
chr2_+_60962228
|
0.538
|
NM_002908
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chrX_+_24393264
|
0.537
|
NM_001142386
NM_005391
|
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3
|
|
chr20_+_36867708
|
0.537
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr2_-_153282104
|
0.535
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr9_-_14683459
|
0.535
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr1_+_205561709
|
0.533
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr18_+_17575275
|
0.530
|
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr3_-_156904690
|
0.529
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr10_+_76541351
|
0.529
|
NM_001174156
NM_144660
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr2_-_104834344
|
0.525
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr19_+_2818332
|
0.525
|
NM_024967
|
ZNF556
|
zinc finger protein 556
|
|
chr6_-_127706384
|
0.518
|
NM_001002030
NM_001105544
NM_001105545
NM_018479
|
ECHDC1
|
enoyl CoA hydratase domain containing 1
|
|
chr16_+_29582065
|
0.518
|
|
SPN
|
sialophorin
|
|
chr12_+_32003553
|
0.516
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr20_-_39199936
|
0.514
|
|
|
|
|
chr2_-_55413253
|
0.514
|
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr20_+_45564063
|
0.514
|
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr1_-_151333624
|
0.513
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr2_+_113479919
|
0.510
|
NM_014440
|
IL1F6
|
interleukin 1 family, member 6 (epsilon)
|
|
chr8_+_32905027
|
0.510
|
|
|
|
|
chr8_-_27171814
|
0.507
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr14_+_23008723
|
0.505
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr21_+_29593098
|
0.504
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr19_-_6621078
|
0.504
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr21_+_29593088
|
0.504
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr7_+_130445394
|
0.502
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr4_-_68093829
|
0.501
|
NM_001812
|
CENPC1
|
centromere protein C 1
|
|
chr12_+_32003648
|
0.501
|
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr6_-_119297851
|
0.497
|
|
MCM9
|
minichromosome maintenance complex component 9
|
|
chr16_+_55963899
|
0.497
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr16_+_64958025
|
0.496
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr8_+_86538307
|
0.492
|
NM_005181
|
CA3
|
carbonic anhydrase III, muscle specific
|
|
chr1_+_195055483
|
0.489
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr4_-_68093784
|
0.487
|
|
CENPC1
|
centromere protein C 1
|
|
chrX_-_118711144
|
0.487
|
|
SEPT6
|
septin 6
|
|
chr1_+_26744929
|
0.486
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr6_-_32665409
|
0.486
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr7_+_150065594
|
0.484
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr5_+_75414994
|
0.484
|
NM_014979
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
|
chr14_+_30098079
|
0.482
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr2_+_63669596
|
0.480
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr4_+_41632125
|
0.479
|
|
TMEM33
|
transmembrane protein 33
|
|
chr16_+_51722285
|
0.477
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr12_+_67426225
|
0.476
|
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr10_+_96295511
|
0.473
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr21_+_29592984
|
0.472
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_+_60962206
|
0.472
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr1_+_160002746
|
0.472
|
|
ATF6
|
activating transcription factor 6
|
|
chrX_+_115481774
|
0.470
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr16_-_16224791
|
0.470
|
NM_001079528
NM_001171
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr10_-_82039158
|
0.469
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chrX_-_31194867
|
0.468
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr20_+_56318245
|
0.468
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr13_+_59869427
|
0.468
|
NM_001146070
|
TDRD3
|
tudor domain containing 3
|
|
chr6_+_32713160
|
0.467
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr20_+_44180355
|
0.466
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr2_+_187163232
|
0.465
|
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr20_+_42463279
|
0.463
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chrX_+_144707022
|
0.463
|
NM_001144003
NM_001144004
NM_001144005
NM_032539
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr17_-_6958282
|
0.461
|
NM_080914
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chrX_-_55037235
|
0.461
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr14_+_34521854
|
0.461
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr6_+_160068135
|
0.458
|
NM_004906
NM_152858
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr20_+_5679032
|
0.455
|
NM_152504
|
C20orf196
|
chromosome 20 open reading frame 196
|
|
chr9_-_74735669
|
0.455
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr20_+_56318155
|
0.454
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr10_-_120928266
|
0.454
|
|
PRDX3
|
peroxiredoxin 3
|
|
chr5_-_60031695
|
0.451
|
NM_001145208
NM_018369
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr9_-_27563444
|
0.450
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr20_+_47328575
|
0.449
|
|
NCRNA00275
|
non-protein coding RNA 275
|
|
chrX_+_15428820
|
0.449
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr10_-_32384914
|
0.449
|
|
|
|
|
chr6_-_127705244
|
0.449
|
NM_001139510
|
ECHDC1
|
enoyl CoA hydratase domain containing 1
|
|
chr13_+_25726755
|
0.448
|
NM_001260
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr1_-_245308649
|
0.448
|
|
ZNF670-ZNF695
ZNF670
|
ZNF670-ZNF695 read-through transcript
zinc finger protein 670
|
|
chr2_+_219141761
|
0.448
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr14_+_34521945
|
0.447
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr6_+_11202251
|
0.447
|
NM_001135575
|
LOC221710
|
hypothetical protein LOC221710
|
|
chr7_-_104949787
|
0.445
|
NM_019042
|
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae)
|
|
chr18_+_17575542
|
0.444
|
NM_020774
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr4_+_38722593
|
0.444
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr2_+_160277273
|
0.443
|
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr6_-_165995510
|
0.442
|
NM_001130690
NM_006661
|
PDE10A
|
phosphodiesterase 10A
|
|
chr6_-_149847718
|
0.442
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr20_+_52258023
|
0.442
|
|
PFDN4
|
prefoldin subunit 4
|
|
chr14_+_64040979
|
0.441
|
NM_001123329
NM_014950
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr12_-_97562836
|
0.440
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr6_+_116528663
|
0.440
|
NM_152729
|
NT5DC1
|
5'-nucleotidase domain containing 1
|
|
chr12_+_30970625
|
0.438
|
|
|
|
|
chr8_-_143820825
|
0.438
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr1_-_31003181
|
0.437
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr2_+_32244406
|
0.437
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr6_+_131190237
|
0.436
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr9_-_88087192
|
0.433
|
NM_030940
|
ISCA1
|
iron-sulfur cluster assembly 1 homolog (S. cerevisiae)
|
|
chr2_+_208103001
|
0.432
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr4_-_35922288
|
0.432
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr15_-_74963246
|
0.431
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr1_+_205561632
|
0.431
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr22_+_23153529
|
0.430
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr15_+_39839866
|
0.429
|
|
MGA
|
MAX gene associated
|
|
chr2_-_238776932
|
0.429
|
|
ILKAP
|
integrin-linked kinase-associated serine/threonine phosphatase
|
|
chr8_+_22266256
|
0.428
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr4_+_57468847
|
0.426
|
|
REST
|
RE1-silencing transcription factor
|
|
chr5_-_60031654
|
0.426
|
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr21_-_15358996
|
0.425
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chrX_-_118711088
|
0.424
|
|
SEPT6
|
septin 6
|
|
chr3_+_194793691
|
0.423
|
|
OPA1
|
optic atrophy 1 (autosomal dominant)
|
|
chr11_-_107234733
|
0.423
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr8_-_117837240
|
0.422
|
NM_003756
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H
|
|
chr5_+_96297529
|
0.422
|
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr2_+_187162969
|
0.420
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr20_+_52257800
|
0.420
|
NM_002623
|
PFDN4
|
prefoldin subunit 4
|
|
chr10_-_120928327
|
0.420
|
NM_006793
NM_014098
|
PRDX3
|
peroxiredoxin 3
|
|
chr1_+_26744893
|
0.420
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr9_-_15500209
|
0.420
|
NM_021144
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr8_-_33450172
|
0.418
|
NM_032664
|
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase)
|
|
chr19_+_56990222
|
0.418
|
NM_002030
|
FPR3
|
formyl peptide receptor 3
|