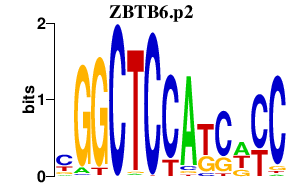
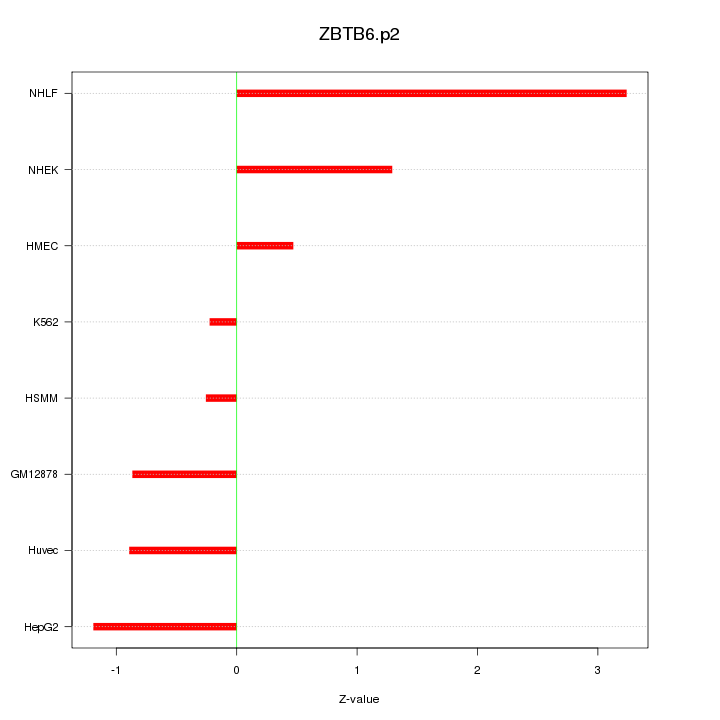
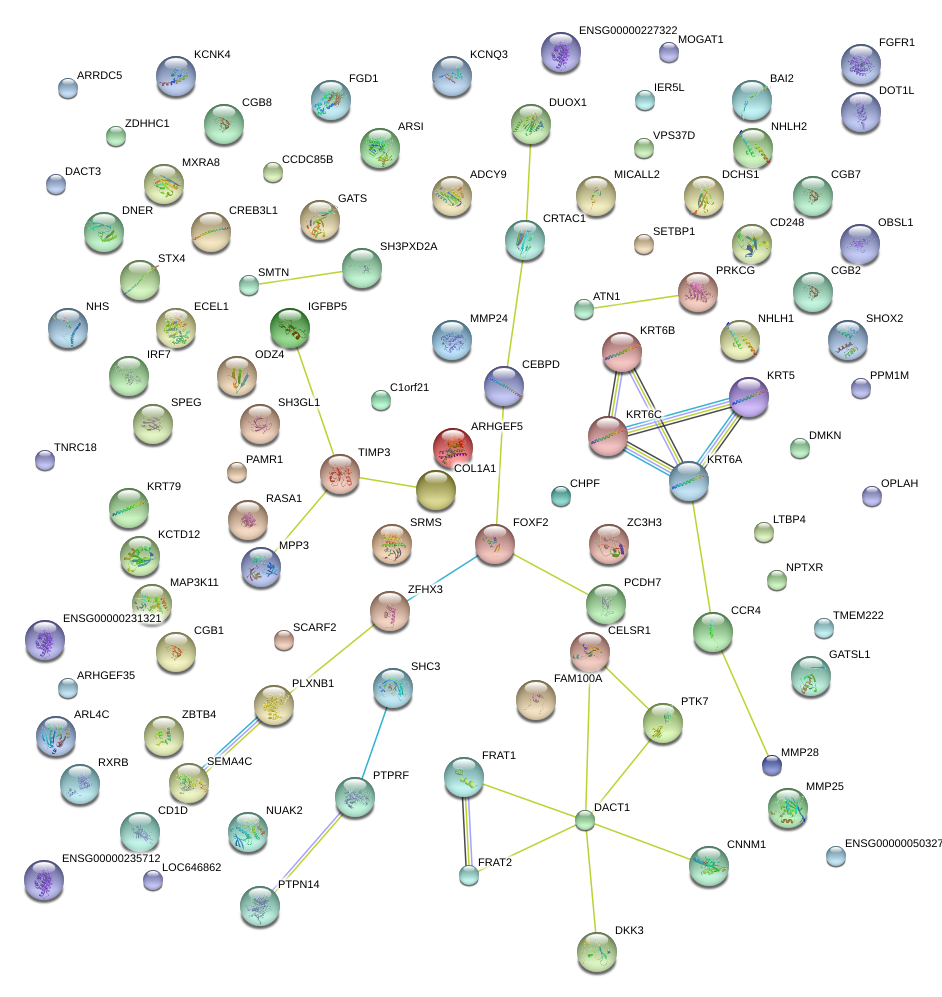
|
chr5_-_149662640
|
1.581
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr12_-_51200347
|
1.430
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr11_-_78829342
|
1.379
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr2_+_220017617
|
1.239
|
|
SPEG
|
SPEG complex locus
|
|
chr11_-_11987414
|
0.992
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr1_+_43769065
|
0.972
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr19_-_40693176
|
0.944
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr3_-_62835601
|
0.927
|
|
|
|
|
chr22_-_37569962
|
0.918
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr12_-_51132129
|
0.856
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr11_-_11987458
|
0.852
|
NM_013253
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr6_+_43151922
|
0.846
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr7_-_1465487
|
0.827
|
|
MICALL2
|
MICAL-like 2
|
|
chr19_+_2115119
|
0.766
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr7_+_143683365
|
0.721
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr15_+_43209422
|
0.720
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr4_+_30331289
|
0.708
|
|
PCDH7
|
protocadherin 7
|
|
chr1_+_116181927
|
0.669
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr1_-_226530350
|
0.664
|
|
|
|
|
chr4_+_30331543
|
0.658
|
|
PCDH7
|
protocadherin 7
|
|
chr19_+_59077241
|
0.657
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr2_-_233060774
|
0.649
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr16_-_66007650
|
0.638
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr11_-_11987115
|
0.634
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr10_-_99780574
|
0.632
|
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr12_-_51173238
|
0.632
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr8_-_144694722
|
0.627
|
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr10_+_101078784
|
0.625
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr11_-_11987333
|
0.624
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr22_+_31527744
|
0.620
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr2_-_217267742
|
0.614
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr22_+_31527678
|
0.608
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr1_+_182622850
|
0.607
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr10_-_105604942
|
0.605
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr3_-_48445719
|
0.605
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr11_+_75056343
|
0.599
|
|
|
|
|
chr11_-_11987198
|
0.592
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr2_-_235070431
|
0.591
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_-_7323667
|
0.587
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr16_-_4105703
|
0.587
|
|
ADCY9
|
adenylate cyclase 9
|
|
chrX_-_50230329
|
0.581
|
NM_001013742
|
DGKK
|
diacylglycerol kinase, kappa
|
|
chr1_-_32002222
|
0.578
|
NM_001703
|
BAI2
|
brain-specific angiogenesis inhibitor 2
|
|
chr11_+_46256161
|
0.577
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr22_-_19122047
|
0.572
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr6_-_33276416
|
0.570
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr16_-_4106022
|
0.568
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr11_+_46255737
|
0.566
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr1_-_1283770
|
0.557
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr16_-_4604895
|
0.555
|
NM_145253
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr14_+_58174489
|
0.551
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr8_-_48813256
|
0.550
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr2_-_96899396
|
0.541
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr11_-_605998
|
0.536
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr7_+_72720093
|
0.530
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr11_-_605920
|
0.525
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr11_+_65414450
|
0.521
|
NM_006848
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr10_+_99069007
|
0.517
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr11_-_35503720
|
0.517
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr9_-_130980359
|
0.513
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr2_-_220116508
|
0.511
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr2_-_230287442
|
0.507
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr16_+_30952382
|
0.505
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chrX_+_49531209
|
0.505
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chrX_+_17303798
|
0.501
|
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr6_+_1335067
|
0.499
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr11_-_65841077
|
0.498
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr11_-_605649
|
0.497
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr8_-_38444982
|
0.491
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr9_-_90983195
|
0.490
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr1_+_156417302
|
0.489
|
|
CD1D
|
CD1d molecule
|
|
chr10_+_22580992
|
0.487
|
|
|
|
|
chr9_-_130980302
|
0.487
|
|
IER5L
|
immediate early response 5-like
|
|
chr11_+_65414677
|
0.480
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr11_+_46256130
|
0.479
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr1_-_203557379
|
0.479
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr11_+_63815348
|
0.478
|
NM_033310
|
KCNK4
|
potassium channel, subfamily K, member 4
|
|
chr11_-_6633427
|
0.478
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr7_-_5429702
|
0.475
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr19_+_45811316
|
0.471
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr22_-_45311730
|
0.467
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr3_+_52254782
|
0.463
|
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M
|
|
chr22_+_29819343
|
0.463
|
|
SMTN
|
smoothelin
|
|
chr19_+_63599268
|
0.463
|
NM_001195135
|
LOC646862
|
hypothetical protein LOC646862
|
|
chr12_+_6907646
|
0.462
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr17_-_45620911
|
0.461
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_+_182622987
|
0.460
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr2_+_223244606
|
0.459
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr20_-_61649259
|
0.455
|
NM_080823
|
SRMS
|
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites
|
|
chr17_-_7323528
|
0.454
|
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr17_-_39265705
|
0.454
|
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr11_-_65139277
|
0.453
|
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr1_-_212791188
|
0.451
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr2_-_220144254
|
0.450
|
|
OBSL1
|
obscurin-like 1
|
|
chrX_+_17303727
|
0.449
|
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr8_-_145187553
|
0.447
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr2_-_220116062
|
0.445
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr7_+_74017015
|
0.441
|
NM_001145063
|
GATSL1
GATSL2
|
GATS protein-like 1
GATS protein-like 2
|
|
chr17_-_45633986
|
0.439
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr16_-_4604781
|
0.438
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr18_+_40513889
|
0.437
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr19_-_4853878
|
0.437
|
NM_001080523
|
ARRDC5
|
arrestin domain containing 5
|
|
chr16_+_3036682
|
0.436
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr19_-_4351470
|
0.435
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chrX_-_54538591
|
0.434
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr13_-_76358370
|
0.433
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr3_-_159306532
|
0.432
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr16_-_71650034
|
0.431
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_-_133561738
|
0.431
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr10_-_99084254
|
0.428
|
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr1_+_27521308
|
0.427
|
|
TMEM222
|
transmembrane protein 222
|
|
chr19_-_54231884
|
0.426
|
NM_033377
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chr14_-_102045849
|
0.425
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr17_+_7549138
|
0.424
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr7_+_72720256
|
0.423
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr8_-_38444675
|
0.419
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr16_-_4605003
|
0.417
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr16_+_65754788
|
0.417
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr5_+_140154627
|
0.411
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr19_+_45915122
|
0.404
|
|
ITPKC
|
inositol 1,4,5-trisphosphate 3-kinase C
|
|
chr11_-_62753574
|
0.404
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr19_+_59333232
|
0.402
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr11_+_129823667
|
0.401
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr1_+_224803123
|
0.399
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr6_-_37773609
|
0.397
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr20_+_33750645
|
0.397
|
NM_080748
|
ROMO1
|
reactive oxygen species modulator 1
|
|
chr11_+_110916442
|
0.395
|
NM_178834
|
LAYN
|
layilin
|
|
chr11_+_63510478
|
0.395
|
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1
|
|
chr10_-_103869923
|
0.395
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr17_-_45633873
|
0.394
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr3_-_75566781
|
0.391
|
|
FAM86DP
|
family with sequence similarity 86, member D, pseudogene
|
|
chr2_-_128138435
|
0.389
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr11_+_63510403
|
0.388
|
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1
|
|
chr16_-_65838813
|
0.384
|
|
FHOD1
|
formin homology 2 domain containing 1
|
|
chr11_-_11986704
|
0.383
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr8_-_144731646
|
0.383
|
NM_145201
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1
|
|
chr5_+_135392482
|
0.382
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr2_+_26768894
|
0.381
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr8_+_68418622
|
0.381
|
|
|
|
|
chr19_-_11477395
|
0.380
|
|
ZNF653
|
zinc finger protein 653
|
|
chr14_+_73073570
|
0.379
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr16_-_65838894
|
0.379
|
|
FHOD1
|
formin homology 2 domain containing 1
|
|
chr12_-_51253875
|
0.378
|
NM_175053
|
KRT74
|
keratin 74
|
|
chr10_-_76829857
|
0.375
|
|
ZNF503
|
zinc finger protein 503
|
|
chr12_+_46133
|
0.374
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr11_-_3104413
|
0.374
|
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr8_+_104582151
|
0.373
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr14_+_93710401
|
0.372
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr12_+_48737753
|
0.371
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr7_-_143622188
|
0.371
|
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr17_+_37942034
|
0.371
|
|
|
|
|
chr11_+_63808858
|
0.370
|
NM_001177358
NM_001170881
|
GPR137
|
G protein-coupled receptor 137
|
|
chr19_+_7801110
|
0.369
|
NM_145245
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chrX_+_12066505
|
0.368
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr4_-_916101
|
0.366
|
|
GAK
|
cyclin G associated kinase
|
|
chr1_+_182622751
|
0.365
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr13_-_20533703
|
0.365
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr16_+_54782751
|
0.365
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr1_-_59020875
|
0.364
|
|
JUN
|
jun proto-oncogene
|
|
chr6_-_2916980
|
0.362
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr14_+_104852225
|
0.362
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr19_-_4351357
|
0.361
|
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr16_+_1696162
|
0.360
|
NM_001040439
NM_015133
|
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr5_+_17270781
|
0.359
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr17_+_37941715
|
0.357
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr19_+_60816770
|
0.356
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chr19_-_14447163
|
0.355
|
NM_000955
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr18_-_43821491
|
0.354
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr9_-_129679760
|
0.353
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr16_+_31619415
|
0.353
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr11_-_64133908
|
0.353
|
|
|
|
|
chr11_-_64368616
|
0.352
|
NM_017525
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr7_+_75928907
|
0.346
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr16_-_30929217
|
0.346
|
NM_052874
|
STX1B
|
syntaxin 1B
|
|
chr10_+_75343352
|
0.345
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr12_-_51153788
|
0.344
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr17_-_39796649
|
0.344
|
NM_198475
|
FAM171A2
|
family with sequence similarity 171, member A2
|
|
chr19_+_1055614
|
0.343
|
NM_001039848
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr19_+_63747638
|
0.343
|
NM_005762
|
TRIM28
|
tripartite motif containing 28
|
|
chr9_-_34448477
|
0.341
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184944
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr9_-_21549696
|
0.341
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr19_+_53559468
|
0.341
|
NM_012451
|
SYNGR4
|
synaptogyrin 4
|
|
chr17_+_45965044
|
0.340
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr4_+_30330963
|
0.339
|
NM_001173523
NM_002589
NM_032456
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr17_-_39437362
|
0.338
|
NM_004160
|
PYY
|
peptide YY
|
|
chr10_-_73203202
|
0.337
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr19_+_45799060
|
0.336
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr19_-_17306538
|
0.335
|
NM_020959
|
ANO8
|
anoctamin 8
|
|
chr17_+_3993166
|
0.332
|
NM_144611
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr10_+_72642274
|
0.331
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr15_+_39573363
|
0.331
|
NM_002220
|
ITPKA
|
inositol 1,4,5-trisphosphate 3-kinase A
|
|
chr8_-_144731498
|
0.330
|
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1
|
|
chr18_-_33399601
|
0.329
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr6_-_33276426
|
0.328
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr19_+_63747829
|
0.326
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr5_+_135392624
|
0.326
|
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr17_+_37941790
|
0.326
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr16_-_85146181
|
0.326
|
NM_001159380
NM_001159377
NM_001159378
NM_001159379
NM_022764
|
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing
|
|
chr3_-_45162901
|
0.325
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr16_+_30952350
|
0.324
|
|
STX4
|
syntaxin 4
|
|
chr6_-_39398293
|
0.324
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|