|
chr16_+_85169615
|
1.823
|
NM_005250
|
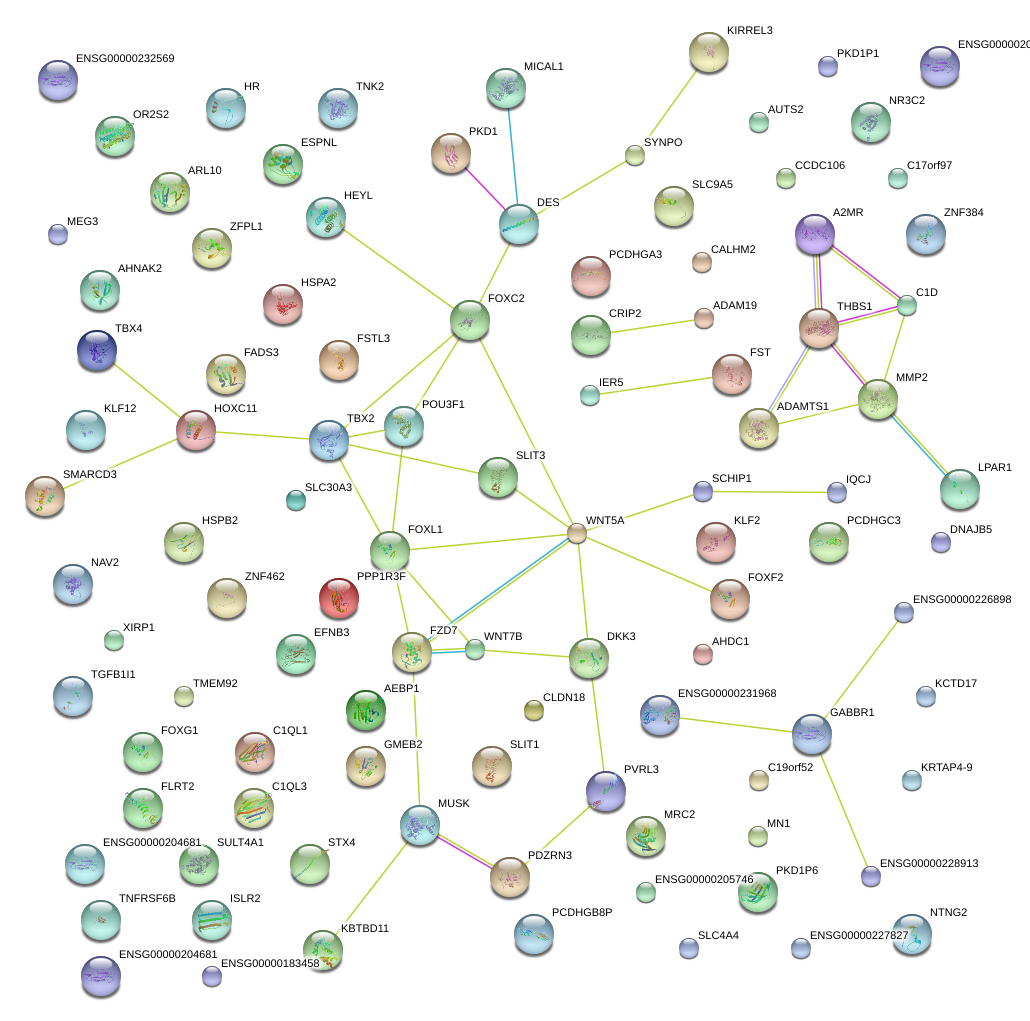
FOXL1
|
forkhead box L1
|
|
chr3_-_73756668
|
1.786
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr9_-_112840064
|
1.375
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr4_-_149582948
|
1.267
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr17_-_40594652
|
1.185
|
|
|
|
|
chr4_-_149582754
|
1.175
|
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr4_+_72271218
|
1.154
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_-_156935283
|
1.116
|
|
|
|
|
chr22_-_26527469
|
1.036
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr7_+_44110527
|
1.025
|
|
AEBP1
|
AE binding protein 1
|
|
chr7_+_44110470
|
0.959
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr3_+_139211639
|
0.935
|
NM_016369
|
CLDN18
|
claudin 18
|
|
chr14_+_28306633
|
0.902
|
|
FOXG1
|
forkhead box G1
|
|
chr21_-_27139143
|
0.870
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr21_-_27139563
|
0.855
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr11_-_126375864
|
0.850
|
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr5_-_168660293
|
0.842
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr1_-_39877928
|
0.840
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr3_-_55490401
|
0.838
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr14_+_100362204
|
0.815
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr22_+_35777561
|
0.799
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr19_+_16296632
|
0.788
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_+_111288669
|
0.781
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr17_+_58058783
|
0.770
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr5_+_150008030
|
0.766
|
|
SYNPO
|
synaptopodin
|
|
chr6_-_169392771
|
0.766
|
|
|
|
|
chr9_-_35948150
|
0.760
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr1_-_38285033
|
0.758
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr17_+_45703765
|
0.757
|
NM_001168215
|
TMEM92
|
transmembrane protein 92
|
|
chr7_+_27190688
|
0.731
|
|
|
|
|
chr17_+_56832492
|
0.727
|
|
TBX2
|
T-box 2
|
|
chr9_+_108665168
|
0.724
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr17_+_56884560
|
0.712
|
|
TBX4
|
T-box 4
|
|
chr9_+_112470871
|
0.712
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr12_-_6668880
|
0.709
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr5_-_156935317
|
0.708
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr3_-_39209075
|
0.703
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr12_+_52653176
|
0.702
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr3_+_160964574
|
0.699
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_75056343
|
0.696
|
|
|
|
|
chr2_-_27339278
|
0.692
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr3_-_197120276
|
0.690
|
NM_005781
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr2_+_219991342
|
0.683
|
NM_001927
|
DES
|
desmin
|
|
chr16_+_31390567
|
0.681
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr14_+_64076916
|
0.679
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr20_-_61728774
|
0.676
|
NM_012384
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr6_-_29703913
|
0.674
|
NM_021903
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr14_+_64077171
|
0.674
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr16_+_30951915
|
0.670
|
|
STX4
|
syntaxin 4
|
|
chr22_-_42589543
|
0.655
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr1_-_27802947
|
0.650
|
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr16_+_54070302
|
0.646
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr9_+_134027154
|
0.642
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr17_+_260408
|
0.639
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr6_+_1335067
|
0.634
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr1_-_39877883
|
0.631
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr11_-_126375895
|
0.622
|
NM_001161707
NM_032531
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr5_+_140835788
|
0.621
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr11_-_11987115
|
0.617
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr19_+_60851207
|
0.614
|
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr17_-_40401138
|
0.613
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr19_+_10900502
|
0.613
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr14_-_104515738
|
0.611
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr5_+_140835727
|
0.608
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr17_+_36515166
|
0.600
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr17_+_7549138
|
0.599
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr2_+_202607554
|
0.599
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr7_+_68702252
|
0.588
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr3_+_112273464
|
0.588
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr15_+_37660568
|
0.583
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr19_+_16296734
|
0.581
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr9_+_34980266
|
0.579
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chrX_+_49013424
|
0.573
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr8_+_1909437
|
0.573
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr16_+_65840275
|
0.565
|
NM_004594
|
SLC9A5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr5_+_52812165
|
0.564
|
|
FST
|
follistatin
|
|
chr17_-_40674883
|
0.563
|
|
|
|
|
chr2_+_238673689
|
0.561
|
NM_194312
|
ESPNL
|
espin-like
|
|
chr11_+_64608307
|
0.560
|
|
ZFPL1
|
zinc finger protein-like 1
|
|
chr1_+_179324458
|
0.552
|
|
IER5
|
immediate early response 5
|
|
chr9_-_112840801
|
0.539
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr9_+_34979674
|
0.539
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr22_-_44751671
|
0.538
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr16_+_85158357
|
0.531
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr19_+_627346
|
0.531
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr5_+_175725050
|
0.529
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr14_+_100362236
|
0.526
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr8_-_22044463
|
0.522
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr20_-_56859352
|
0.521
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr7_-_150576660
|
0.520
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr6_-_109883864
|
0.517
|
NM_001159291
NM_022765
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr13_-_73605914
|
0.516
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr10_-_105202012
|
0.515
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr16_-_2099854
|
0.514
|
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr20_+_61798446
|
0.510
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr14_+_105012106
|
0.509
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr1_-_234096793
|
0.508
|
|
|
|
|
chr11_-_61415429
|
0.507
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr14_+_85066265
|
0.506
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr11_-_11986704
|
0.505
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr12_+_55808514
|
0.504
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr11_+_19691397
|
0.503
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr15_+_72209039
|
0.497
|
NM_001130137
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr1_+_168899936
|
0.493
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr1_-_1233109
|
0.493
|
NM_030649
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3
|
|
chr1_-_40140126
|
0.493
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr19_+_60487345
|
0.492
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr12_-_50871953
|
0.491
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr16_-_1999760
|
0.487
|
NM_178167
|
ZNF598
|
zinc finger protein 598
|
|
chr15_+_72208751
|
0.487
|
NM_001130136
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr11_+_131285747
|
0.485
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr11_-_11987198
|
0.484
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr3_+_128874458
|
0.483
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr6_+_45498262
|
0.483
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr19_-_50987625
|
0.483
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr19_-_54835162
|
0.481
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr3_+_50272648
|
0.481
|
|
|
|
|
chr17_-_28644118
|
0.481
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr2_-_26058831
|
0.480
|
|
KIF3C
|
kinesin family member 3C
|
|
chr7_-_6557591
|
0.479
|
NM_001145118
|
GRID2IP
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein
|
|
chr13_-_109236897
|
0.476
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr17_-_15185623
|
0.474
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr19_-_53912025
|
0.474
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr12_-_3732614
|
0.473
|
NM_001144958
NM_001144959
NM_032680
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr11_-_11600118
|
0.472
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr17_+_7252225
|
0.472
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr11_-_66898223
|
0.471
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chrX_-_53367246
|
0.470
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chrX_+_101792949
|
0.467
|
NM_001099410
NM_001099411
NM_014710
NM_001184727
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr17_+_73738985
|
0.464
|
NM_001145529
|
TMEM235
|
transmembrane protein 235
|
|
chr1_+_155130146
|
0.463
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr8_-_116750427
|
0.462
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr6_+_45498354
|
0.459
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr22_-_17545872
|
0.457
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr2_-_27967458
|
0.453
|
|
LOC100302650
|
hypothetical LOC100302650
|
|
chr9_+_101623957
|
0.453
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr17_-_44010543
|
0.451
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chrX_+_51944658
|
0.450
|
NM_001098800
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr12_+_6179736
|
0.450
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr11_-_110917632
|
0.449
|
|
|
|
|
chr9_-_130912728
|
0.447
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr16_+_30952350
|
0.447
|
|
STX4
|
syntaxin 4
|
|
chr11_+_65802221
|
0.445
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr11_-_111287657
|
0.444
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr11_-_65396775
|
0.442
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr10_+_72102563
|
0.439
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr3_+_112273352
|
0.437
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr1_+_99502655
|
0.436
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr11_+_19328846
|
0.436
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chrX_-_100801472
|
0.434
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr11_+_111288686
|
0.433
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr2_-_144991386
|
0.432
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_+_87021379
|
0.430
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr19_-_5291700
|
0.429
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr3_+_139200347
|
0.429
|
NM_001002026
|
CLDN18
|
claudin 18
|
|
chr10_+_75343352
|
0.428
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr7_+_43118721
|
0.428
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr1_-_27802688
|
0.428
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr14_+_102662416
|
0.428
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr4_-_140696740
|
0.426
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr9_-_103289154
|
0.424
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr19_+_60487578
|
0.424
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr11_-_35503720
|
0.424
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chrX_+_149282097
|
0.422
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr19_+_7505037
|
0.422
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr1_-_35143570
|
0.422
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr11_-_118055564
|
0.422
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr12_-_6668680
|
0.421
|
|
ZNF384
|
zinc finger protein 384
|
|
chr22_-_17546234
|
0.418
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr2_-_26058941
|
0.417
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr19_-_45611110
|
0.417
|
NM_020956
NM_181882
|
PRX
|
periaxin
|
|
chr17_+_23722689
|
0.417
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr7_-_150576634
|
0.415
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr7_+_100037817
|
0.415
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr12_+_55808696
|
0.415
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr20_+_25176698
|
0.414
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr12_-_70952572
|
0.414
|
|
LOC283392
|
hypothetical LOC283392
|
|
chr5_-_83715947
|
0.414
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr7_+_68701704
|
0.413
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr7_-_27180398
|
0.412
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr16_+_3345889
|
0.412
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr6_+_149109841
|
0.410
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr6_-_29703725
|
0.410
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chrX_-_30237403
|
0.408
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr15_+_41597097
|
0.408
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr11_+_112337491
|
0.407
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr19_-_7896974
|
0.407
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr3_-_9570412
|
0.406
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr8_-_124613557
|
0.405
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr1_-_200946038
|
0.405
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr1_+_99502435
|
0.402
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr17_+_7846636
|
0.401
|
NM_000180
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific)
|
|
chrX_-_107866262
|
0.401
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr15_+_72315719
|
0.399
|
NM_025055
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr7_-_135063461
|
0.399
|
NM_012450
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporters), member 4
|
|
chr2_+_220033777
|
0.399
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr19_+_482737
|
0.398
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr15_-_40236061
|
0.398
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr16_+_30952382
|
0.396
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr9_-_139235595
|
0.395
|
NM_031297
|
RNF208
|
ring finger protein 208
|