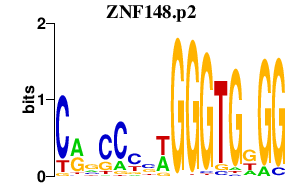
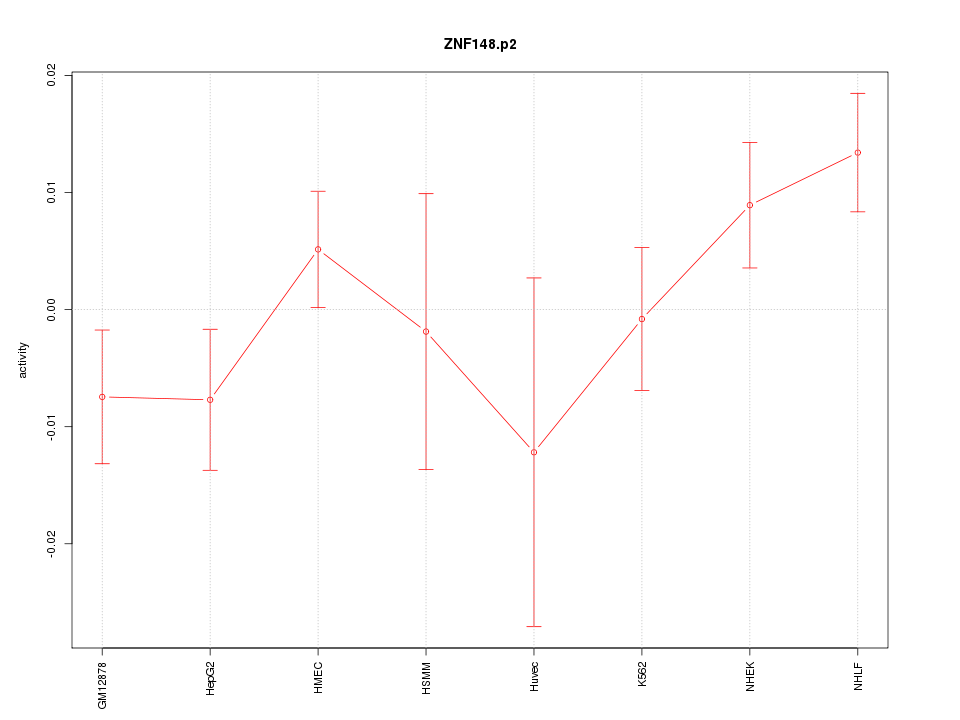
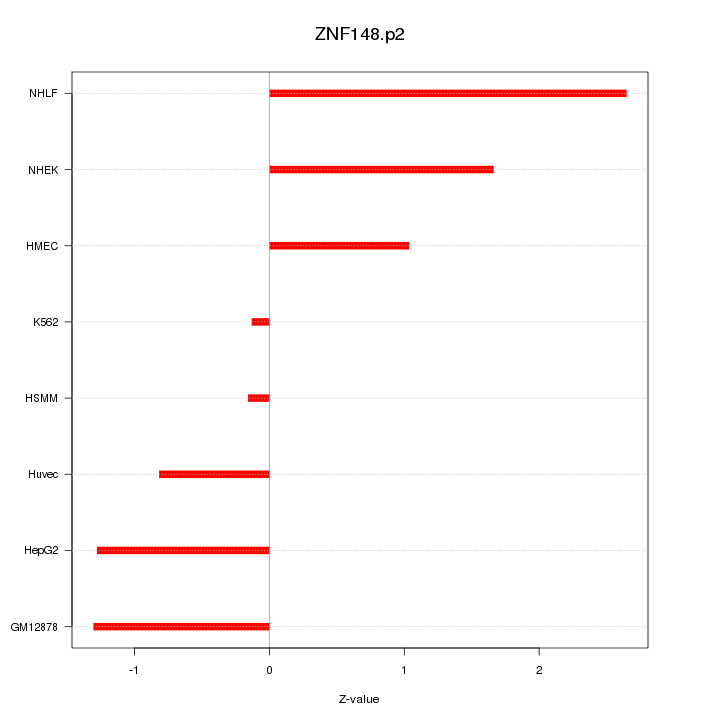
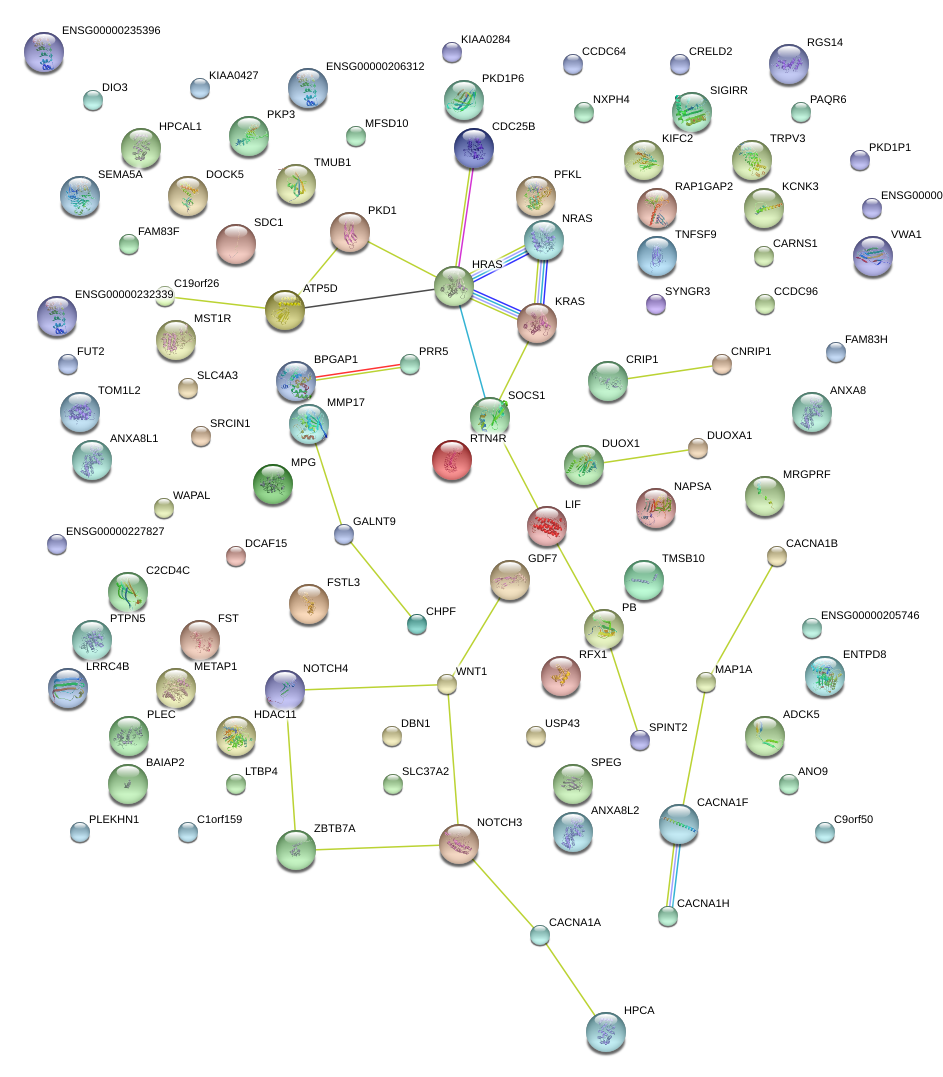
|
chr3_-_49916304
|
1.212
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr19_-_4016467
|
1.122
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr22_+_48698337
|
1.102
|
|
CRELD2
|
cysteine-rich with EGF-like domains 2
|
|
chr2_-_220116508
|
1.029
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr17_+_76623553
|
0.996
|
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr1_+_891739
|
0.974
|
NM_001160184
NM_032129
|
PLEKHN1
|
pleckstrin homology domain containing, family N member 1
|
|
chr19_-_15172773
|
0.925
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr22_+_48698282
|
0.876
|
NM_001135101
NM_024324
|
CRELD2
|
cysteine-rich with EGF-like domains 2
|
|
chr1_-_154484341
|
0.826
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr19_-_55763091
|
0.784
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr11_-_68537243
|
0.741
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr5_-_176833259
|
0.724
|
NM_004395
|
DBN1
|
drebrin 1
|
|
chr15_+_43209422
|
0.687
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr3_+_13496714
|
0.676
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr17_+_76623540
|
0.673
|
NM_001144888
NM_006340
NM_017450
NM_017451
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr11_+_384199
|
0.653
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr19_+_1192767
|
0.643
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chrX_-_134060329
|
0.638
|
|
NCRNA00087
|
non-protein coding RNA 87
|
|
chr2_-_20288647
|
0.633
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr19_+_627346
|
0.623
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr19_+_53891039
|
0.622
|
NM_000511
NM_001097638
|
FUT2
|
fucosyltransferase 2 (secretor status included)
|
|
chr19_+_1192811
|
0.621
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr8_-_144887876
|
0.621
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr8_+_25097983
|
0.612
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr5_+_176726918
|
0.611
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr11_+_384238
|
0.609
|
|
PKP3
|
plakophilin 3
|
|
chr1_-_1041598
|
0.604
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr12_+_130878870
|
0.601
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr17_-_3408038
|
0.596
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr14_+_104402626
|
0.595
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr5_-_176833197
|
0.593
|
|
DBN1
|
drebrin 1
|
|
chr15_-_43209348
|
0.593
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr2_+_20729904
|
0.591
|
NM_182828
|
GDF7
|
growth differentiation factor 7
|
|
chr19_+_13924287
|
0.590
|
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr2_-_20288108
|
0.582
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr22_+_38720881
|
0.580
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr22_+_43476755
|
0.577
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr22_-_28972683
|
0.572
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr2_-_220116062
|
0.566
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr8_+_145662510
|
0.555
|
NM_145754
|
KIFC2
|
kinesin family member C2
|
|
chr16_-_11257539
|
0.546
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr7_-_150411504
|
0.545
|
NM_001136044
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1
|
|
chr14_+_101097440
|
0.543
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr2_+_84986273
|
0.539
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr17_-_34015631
|
0.539
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr2_+_26768894
|
0.537
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr22_-_18635614
|
0.533
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr1_+_33124658
|
0.533
|
NM_002143
|
HPCA
|
hippocalcin
|
|
chr10_+_47875229
|
0.532
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr16_+_68233
|
0.531
|
|
MPG
|
N-methylpurine-DNA glycosylase
|
|
chr14_+_105024576
|
0.530
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr11_-_18769703
|
0.525
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr19_-_1188840
|
0.521
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr16_+_1143738
|
0.516
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr8_+_145568555
|
0.508
|
|
ADCK5
|
aarF domain containing kinase 5
|
|
chr19_+_45807909
|
0.507
|
|
|
|
|
chr5_-_9599157
|
0.506
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr1_-_1041321
|
0.486
|
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr1_+_1360765
|
0.483
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr22_+_44828338
|
0.481
|
|
|
|
|
chr11_+_124438217
|
0.480
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr16_+_1979959
|
0.477
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr2_+_220200530
|
0.476
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr19_+_43446937
|
0.475
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_43447078
|
0.474
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr9_-_131422842
|
0.473
|
NM_199350
|
C9orf50
|
chromosome 9 open reading frame 50
|
|
chrX_-_48976714
|
0.472
|
NM_005183
|
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit
|
|
chr19_+_45794895
|
0.470
|
NM_001042544
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr4_-_7095566
|
0.469
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr2_+_220015016
|
0.468
|
|
SPEG
|
SPEG complex locus
|
|
chr12_+_118911859
|
0.466
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr11_-_525532
|
0.465
|
NM_001130442
NM_005343
NM_176795
|
HRAS
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog
|
|
chr20_+_3724905
|
0.463
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr19_+_6482009
|
0.455
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr17_+_9489578
|
0.454
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr8_-_145090045
|
0.451
|
NM_201382
|
PLEC
|
plectin
|
|
chr19_-_360138
|
0.450
|
NM_001136263
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr1_+_32992131
|
0.447
|
|
|
|
|
chr9_-_139455611
|
0.446
|
NM_001033113
NM_198585
|
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8
|
|
chr12_-_131415854
|
0.439
|
NM_001122636
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr15_+_41597097
|
0.438
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr11_+_66939720
|
0.437
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr15_-_19221463
|
0.435
|
|
|
|
|
chr18_+_44319365
|
0.435
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr11_-_431956
|
0.434
|
|
ANO9
|
anoctamin 9
|
|
chr19_+_1192742
|
0.430
|
NM_001001975
NM_001687
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr8_+_145568526
|
0.430
|
NM_174922
|
ADCK5
|
aarF domain containing kinase 5
|
|
chr12_+_55896844
|
0.428
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr12_+_47658502
|
0.427
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr5_+_52812165
|
0.427
|
|
FST
|
follistatin
|
|
chr9_+_139892049
|
0.426
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr19_+_1192770
|
0.424
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr17_+_2646481
|
0.424
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr19_+_43447262
|
0.423
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr17_-_17816425
|
0.420
|
NM_001033551
NM_001082968
|
TOM1L2
|
target of myb1-like 2 (chicken)
|
|
chr19_-_13978096
|
0.418
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr11_-_407324
|
0.418
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr21_+_44560779
|
0.416
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr4_-_2905649
|
0.415
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr16_-_2125825
|
0.413
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr19_-_43412170
|
0.413
|
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr1_-_212791188
|
0.413
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr19_+_1192799
|
0.410
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr10_-_99780336
|
0.409
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr2_-_127694123
|
0.408
|
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr1_+_182622850
|
0.406
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr20_+_61394982
|
0.406
|
NM_020882
|
COL20A1
|
collagen, type XX, alpha 1
|
|
chr17_-_77421980
|
0.404
|
NM_001185077
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr3_-_57174353
|
0.400
|
|
IL17RD
|
interleukin 17 receptor D
|
|
chr19_-_48977247
|
0.399
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr4_+_2390469
|
0.399
|
NM_001193282
|
LOC402160
|
hypothetical protein LOC402160
|
|
chr11_+_63510403
|
0.399
|
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1
|
|
chr7_+_72720256
|
0.399
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr17_-_77422463
|
0.396
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr8_-_145090892
|
0.393
|
NM_201381
|
PLEC
|
plectin
|
|
chr11_-_75599433
|
0.392
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr8_-_144969529
|
0.392
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr17_+_7846636
|
0.391
|
NM_000180
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific)
|
|
chr15_+_38318528
|
0.390
|
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr19_+_13924318
|
0.390
|
NM_138353
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr18_+_40513889
|
0.387
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr3_-_49178773
|
0.384
|
NM_022903
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr3_-_187025501
|
0.382
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_-_48700824
|
0.379
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr3_-_49178762
|
0.378
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr19_-_14391129
|
0.378
|
|
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
|
|
chr16_+_29724926
|
0.377
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr11_+_47386707
|
0.375
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr17_+_7283412
|
0.375
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr9_+_131974506
|
0.374
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr18_-_26876622
|
0.374
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr11_-_61104866
|
0.373
|
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr19_+_7505723
|
0.372
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr17_-_1335763
|
0.370
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr2_+_234286376
|
0.370
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr7_-_150411345
|
0.369
|
NM_031434
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1
|
|
chr11_+_2279932
|
0.368
|
|
TSPAN32
|
tetraspanin 32
|
|
chr14_-_20636337
|
0.367
|
|
ZNF219
|
zinc finger protein 219
|
|
chr3_-_49882308
|
0.367
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr8_+_120497732
|
0.367
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr15_-_19336666
|
0.367
|
NM_207355
|
POTEB
|
POTE ankyrin domain family, member B
|
|
chr4_-_2906335
|
0.367
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr19_-_43498220
|
0.366
|
NM_001039671
NM_001145461
NM_001145462
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr3_-_49178738
|
0.366
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr19_-_52614280
|
0.365
|
|
MEIS3
|
Meis homeobox 3
|
|
chr11_-_46678736
|
0.365
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr17_+_1892026
|
0.365
|
NM_080822
|
OVCA2
|
ovarian tumor suppressor candidate 2
|
|
chrX_+_152643516
|
0.363
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr11_+_124438407
|
0.363
|
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr19_+_447453
|
0.362
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr20_+_32610161
|
0.359
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr6_-_38715613
|
0.358
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr2_+_10506233
|
0.358
|
|
|
|
|
chr20_-_2729227
|
0.358
|
|
CPXM1
|
carboxypeptidase X (M14 family), member 1
|
|
chr17_-_24972471
|
0.357
|
NM_032854
|
CORO6
|
coronin 6
|
|
chrX_+_139912421
|
0.355
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chr17_+_4683280
|
0.355
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr7_+_73080422
|
0.352
|
|
ELN
|
elastin
|
|
chr9_-_139235595
|
0.352
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr10_+_120957084
|
0.352
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr19_-_50838824
|
0.351
|
NM_001193269
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr2_+_63131466
|
0.350
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr18_-_33399601
|
0.350
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr20_-_49073023
|
0.349
|
NM_002237
|
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1
|
|
chr15_+_29406335
|
0.349
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr10_-_73203202
|
0.348
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr3_-_47595190
|
0.345
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr1_-_22136263
|
0.344
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr19_+_7505590
|
0.344
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr20_-_3714905
|
0.342
|
NM_001810
|
CENPB
|
centromere protein B, 80kDa
|
|
chr7_+_73080387
|
0.341
|
|
ELN
|
elastin
|
|
chr19_-_5291700
|
0.340
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_-_68275520
|
0.339
|
NM_001039656
NM_004923
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr19_-_1989937
|
0.336
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr1_+_182622987
|
0.336
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr7_-_2320576
|
0.335
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr16_+_82782223
|
0.334
|
NM_001145400
NM_139174
|
ADAD2
|
adenosine deaminase domain containing 2
|
|
chr7_+_149733309
|
0.334
|
|
|
|
|
chr19_-_4017739
|
0.333
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr11_-_68275486
|
0.333
|
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr1_-_39910296
|
0.332
|
NM_032526
|
NT5C1A
|
5'-nucleotidase, cytosolic IA
|
|
chr11_+_2355099
|
0.331
|
|
CD81
|
CD81 molecule
|
|
chr11_+_63510876
|
0.331
|
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1
|
|
chr19_-_11477395
|
0.330
|
|
ZNF653
|
zinc finger protein 653
|
|
chr8_+_145137354
|
0.330
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr16_+_29725311
|
0.329
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr19_-_43412152
|
0.329
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr19_+_13066956
|
0.328
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_45728207
|
0.327
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr19_-_48976852
|
0.326
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr22_-_35113947
|
0.326
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr14_-_56342097
|
0.325
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr19_-_18749728
|
0.324
|
|
|
|
|
chr16_+_2068186
|
0.324
|
|
TSC2
|
tuberous sclerosis 2
|
|
chr12_+_48641545
|
0.323
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr1_+_156229686
|
0.321
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr17_+_74531820
|
0.320
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_-_6162669
|
0.319
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr17_-_40565207
|
0.318
|
NM_133373
|
PLCD3
|
phospholipase C, delta 3
|
|
chr19_-_53706644
|
0.317
|
|
|
|