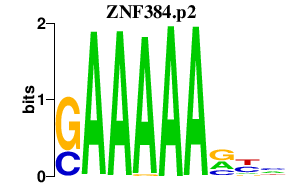
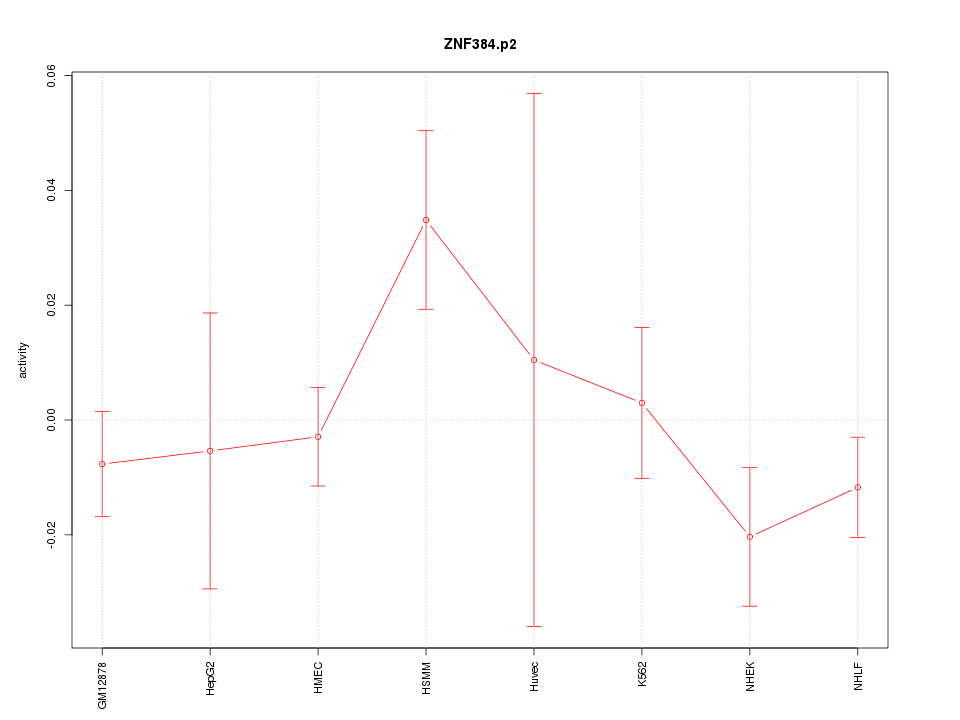
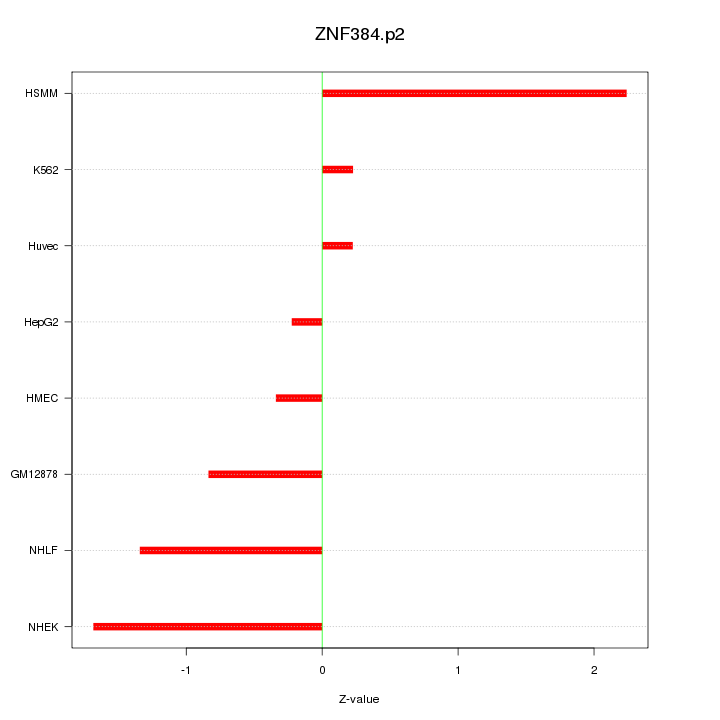
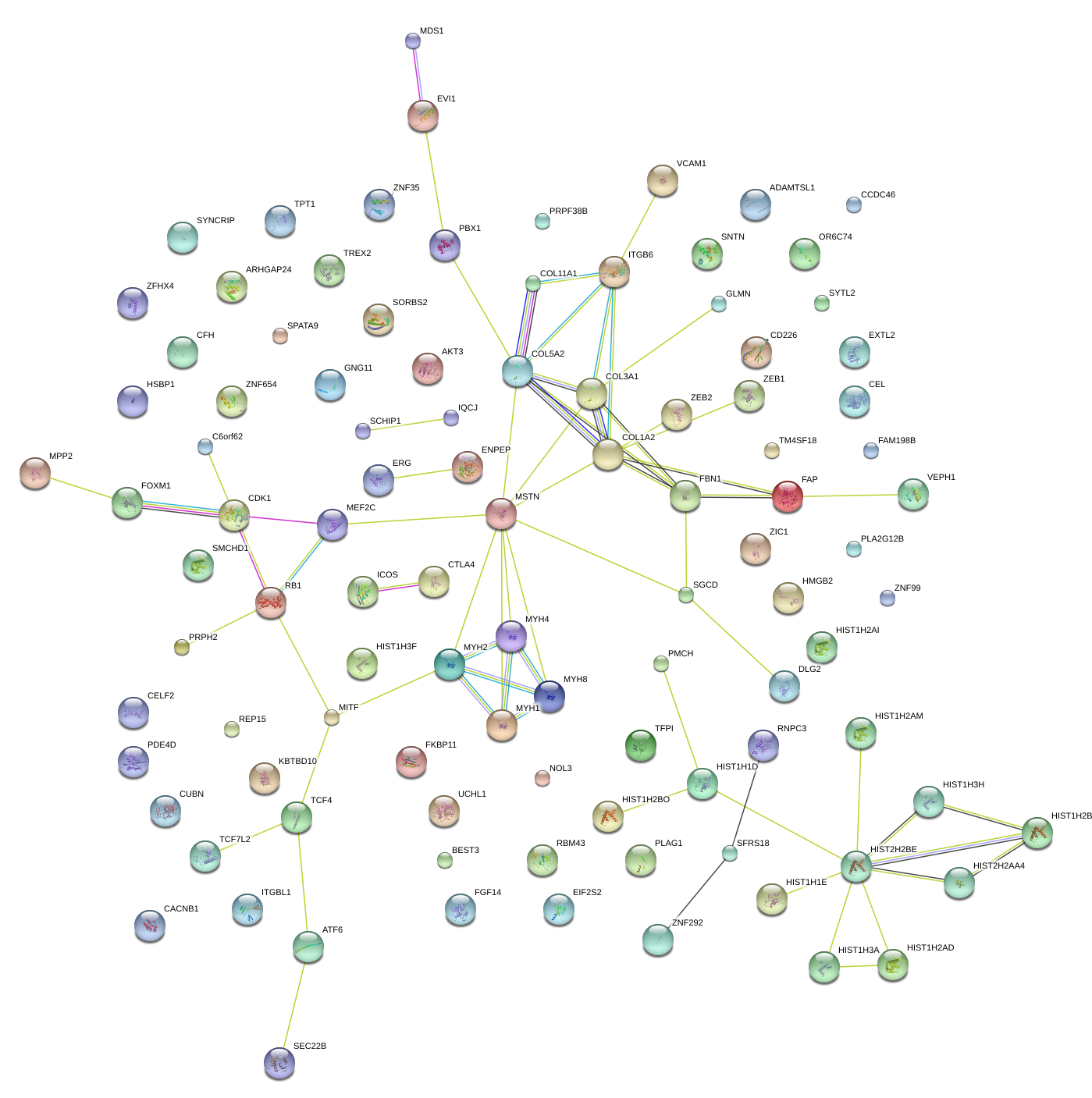
|
chr2_-_144994349
|
3.039
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144994038
|
2.875
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_+_189547323
|
2.474
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_-_24827265
|
2.274
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr2_-_189752575
|
2.252
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_+_155686333
|
2.028
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr2_-_188127430
|
1.962
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr5_-_95044415
|
1.775
|
NM_031952
|
SPATA9
|
spermatogenesis associated 9
|
|
chr13_-_101852028
|
1.728
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr6_-_26343139
|
1.591
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr2_+_189547358
|
1.580
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_+_26359805
|
1.551
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr1_+_194887630
|
1.530
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr5_-_88155360
|
1.529
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_189547377
|
1.526
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr3_+_88270951
|
1.477
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr12_-_101115677
|
1.405
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr6_-_86407875
|
1.377
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_190552744
|
1.341
|
NM_001039152
|
RGS21
|
regulator of G-protein signaling 21
|
|
chr2_+_170074457
|
1.337
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr1_-_242073094
|
1.287
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr13_+_100902942
|
1.256
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr12_+_53927248
|
1.256
|
NM_001005490
|
OR6C74
|
olfactory receptor, family 6, subfamily C, member 74
|
|
chr4_+_87070449
|
1.099
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr18_-_65775139
|
1.093
|
NM_006566
|
CD226
|
CD226 molecule
|
|
chr2_+_204509698
|
1.080
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr17_-_10362583
|
1.058
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr3_-_158695832
|
1.054
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr3_-_150533948
|
1.014
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr7_-_15568099
|
1.012
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr6_-_138581319
|
0.999
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr4_+_111616630
|
0.999
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr12_-_68379331
|
0.988
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr19_-_22744623
|
0.988
|
NM_001080409
|
ZNF99
|
zinc finger protein 99
|
|
chr21_-_38955487
|
0.976
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr8_+_77928372
|
0.964
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr21_-_38792173
|
0.956
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_-_17211795
|
0.946
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr1_-_101132833
|
0.930
|
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr10_+_11246934
|
0.927
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_+_160269734
|
0.917
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr2_-_190635566
|
0.912
|
NM_005259
|
MSTN
|
myostatin
|
|
chr1_-_103346639
|
0.900
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr4_-_159313167
|
0.897
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_+_109043550
|
0.896
|
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr2_-_160765008
|
0.882
|
|
ITGB6
|
integrin, beta 6
|
|
chr5_+_155686750
|
0.875
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr21_+_25856311
|
0.871
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr1_+_162795328
|
0.862
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_170864111
|
0.861
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr4_+_40953698
|
0.854
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr6_-_99955987
|
0.851
|
|
SFRS18
|
splicing factor, arginine/serine-rich 18
|
|
chr13_-_44812246
|
0.846
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr1_+_109043711
|
0.840
|
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr15_-_46725087
|
0.837
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr9_+_18464138
|
0.834
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr6_+_88026338
|
0.826
|
|
ZNF292
|
zinc finger protein 292
|
|
chr11_-_85015961
|
0.820
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr5_-_58918031
|
0.818
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_+_62208256
|
0.818
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr6_+_27883877
|
0.816
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr7_+_93388951
|
0.805
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr13_+_47948586
|
0.803
|
|
RB1
|
retinoblastoma 1
|
|
chr2_-_188127294
|
0.792
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr1_+_143807876
|
0.785
|
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr1_+_100957883
|
0.782
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chrX_-_135783496
|
0.780
|
|
|
|
|
chr3_+_70068440
|
0.771
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr7_+_93862137
|
0.769
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr18_-_51406841
|
0.768
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr2_+_171316462
|
0.767
|
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr2_-_151826504
|
0.766
|
NM_198557
|
RBM43
|
RNA binding motif protein 43
|
|
chr1_+_160002746
|
0.766
|
|
ATF6
|
activating transcription factor 6
|
|
chr2_-_162808123
|
0.758
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr10_-_74384569
|
0.758
|
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr3_+_63613383
|
0.757
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr4_-_186934059
|
0.757
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_159311959
|
0.752
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_174491270
|
0.750
|
NM_001130689
|
HMGB2
|
high-mobility group box 2
|
|
chr11_-_85115105
|
0.744
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr3_+_148609841
|
0.740
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr17_-_61253122
|
0.739
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr12_+_27740694
|
0.736
|
NM_001029874
|
REP15
|
RAB15 effector protein
|
|
chr1_-_58920315
|
0.730
|
|
|
|
|
chr3_+_44667504
|
0.719
|
|
ZNF35
|
zinc finger protein 35
|
|
chr12_+_50342814
|
0.711
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr5_-_95774444
|
0.710
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr9_-_14076901
|
0.710
|
|
NFIB
|
nuclear factor I/B
|
|
chr7_+_18502342
|
0.706
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chrX_+_77889861
|
0.705
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr9_+_88953378
|
0.703
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr1_+_190871868
|
0.702
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr10_+_11247449
|
0.699
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_83771386
|
0.698
|
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr6_-_139654807
|
0.698
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr10_-_61570665
|
0.697
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_+_74462654
|
0.695
|
|
CD109
|
CD109 molecule
|
|
chr2_+_5750229
|
0.694
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr7_+_120415958
|
0.686
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr12_-_90029672
|
0.685
|
NM_002345
|
LUM
|
lumican
|
|
chr11_-_27637771
|
0.682
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr15_-_53440069
|
0.673
|
|
CCPG1
|
cell cycle progression 1
|
|
chr3_+_29297806
|
0.665
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr16_+_12902955
|
0.664
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr11_-_102331643
|
0.662
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr6_+_152053323
|
0.662
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr8_-_13018123
|
0.661
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr2_-_224175386
|
0.652
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr9_-_35948150
|
0.649
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr17_-_10313600
|
0.647
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr6_+_74462577
|
0.647
|
|
CD109
|
CD109 molecule
|
|
chr4_-_159312973
|
0.645
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr6_+_12398514
|
0.642
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr7_+_120416103
|
0.639
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr20_+_9236446
|
0.637
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr8_-_82606104
|
0.628
|
NM_001105281
|
FABP12
|
fatty acid binding protein 12
|
|
chr8_-_82769761
|
0.626
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr1_-_195436258
|
0.625
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr5_+_178419938
|
0.623
|
NM_014594
|
ZNF354C
|
zinc finger protein 354C
|
|
chr12_+_79634820
|
0.615
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr1_+_144152864
|
0.608
|
|
TXNIP
|
thioredoxin interacting protein
|
|
chr2_+_99344060
|
0.601
|
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr5_+_40877053
|
0.589
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr11_-_32772731
|
0.588
|
NM_001008391
|
CCDC73
|
coiled-coil domain containing 73
|
|
chr7_+_123353144
|
0.587
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr7_-_92585205
|
0.581
|
|
SAMD9
|
sterile alpha motif domain containing 9
|
|
chr8_+_72918889
|
0.579
|
|
|
|
|
chr5_-_172016174
|
0.579
|
|
|
|
|
chr12_+_79625538
|
0.573
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chrX_-_153698930
|
0.573
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr12_-_9913199
|
0.572
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr8_-_15140162
|
0.570
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr13_+_31503436
|
0.566
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr1_-_155936904
|
0.565
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr17_-_59825221
|
0.565
|
|
|
|
|
chr7_-_92585253
|
0.564
|
NM_001193307
NM_017654
|
SAMD9
|
sterile alpha motif domain containing 9
|
|
chr5_+_141468393
|
0.563
|
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr7_-_33047042
|
0.558
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr8_-_30826074
|
0.556
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr18_-_21184824
|
0.555
|
|
|
|
|
chr5_-_55564942
|
0.554
|
NM_024669
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chr7_-_37922902
|
0.552
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr4_-_101658094
|
0.552
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr18_-_35634247
|
0.552
|
|
LOC647946
|
hypothetical LOC647946
|
|
chr11_+_65027721
|
0.550
|
|
|
|
|
chr4_+_159311353
|
0.548
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr4_+_123380569
|
0.545
|
|
KIAA1109
|
KIAA1109
|
|
chr17_+_26446082
|
0.540
|
|
NF1
|
neurofibromin 1
|
|
chr5_+_176494690
|
0.540
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chrX_+_100361588
|
0.540
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr7_+_22733626
|
0.539
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr3_-_55496370
|
0.538
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr2_-_17563186
|
0.537
|
NM_001099218
|
RAD51AP2
|
RAD51 associated protein 2
|
|
chr1_+_61315378
|
0.535
|
|
|
|
|
chr10_+_17834256
|
0.533
|
NM_001098844
|
FAM23A
|
family with sequence similarity 23, member A
|
|
chr14_+_55148488
|
0.533
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr10_+_62208094
|
0.530
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr3_-_102888090
|
0.530
|
NM_000986
|
RPL24
|
ribosomal protein L24
|
|
chr15_-_53996597
|
0.526
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr10_-_60792225
|
0.526
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr2_-_210744158
|
0.526
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr7_+_117651947
|
0.525
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr6_-_134680888
|
0.523
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_-_135063461
|
0.522
|
NM_012450
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporters), member 4
|
|
chr13_-_36577800
|
0.521
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr12_-_29828958
|
0.521
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr1_-_79244948
|
0.519
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr15_-_64644758
|
0.513
|
NM_207338
|
LCTL
|
lactase-like
|
|
chr1_+_143807748
|
0.511
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr11_-_57883117
|
0.511
|
NM_001005489
|
OR5B17
|
olfactory receptor, family 5, subfamily B, member 17
|
|
chr1_+_160002733
|
0.510
|
|
ATF6
|
activating transcription factor 6
|
|
chr1_-_171442937
|
0.507
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr13_+_49487390
|
0.506
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr8_-_72436519
|
0.506
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_73167586
|
0.505
|
NM_001168390
|
C10orf105
|
chromosome 10 open reading frame 105
|
|
chr12_-_89976261
|
0.504
|
NM_007035
|
KERA
|
keratocan
|
|
chr5_-_37406643
|
0.504
|
NM_004298
|
NUP155
|
nucleoporin 155kDa
|
|
chr18_+_50139168
|
0.499
|
NM_173529
|
C18orf54
|
chromosome 18 open reading frame 54
|
|
chr8_+_72918772
|
0.499
|
|
LOC100132891
|
hypothetical LOC100132891
|
|
chr13_-_19008884
|
0.499
|
NM_001141968
NM_130785
NM_199254
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2
|
|
chr12_+_91654395
|
0.498
|
NM_001004330
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7
|
|
chr19_+_4720144
|
0.497
|
|
C19orf30
|
chromosome 19 open reading frame 30
|
|
chr16_+_6763810
|
0.496
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_+_8557402
|
0.496
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr8_+_26206648
|
0.495
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr3_-_150993299
|
0.495
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr17_+_26446067
|
0.491
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr11_+_65023038
|
0.490
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr16_-_55042614
|
0.489
|
NM_007006
|
NUDT21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21
|
|
chr11_-_85457736
|
0.489
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr21_-_14840506
|
0.488
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr8_-_27751462
|
0.488
|
|
PBK
|
PDZ binding kinase
|
|
chr1_+_160002693
|
0.488
|
NM_007348
|
ATF6
|
activating transcription factor 6
|
|
chr4_+_170817787
|
0.486
|
|
CLCN3
|
chloride channel 3
|
|
chr12_+_79362256
|
0.485
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr18_-_22699650
|
0.484
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr13_-_47885653
|
0.481
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr2_-_159021201
|
0.478
|
NM_001171637
NM_138803
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr1_-_84928473
|
0.477
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr11_-_85457753
|
0.476
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|