|
chr17_-_35017691
|
2.126
|
NM_006160
|
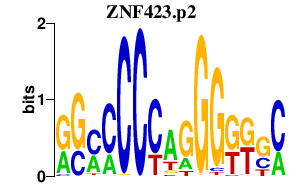
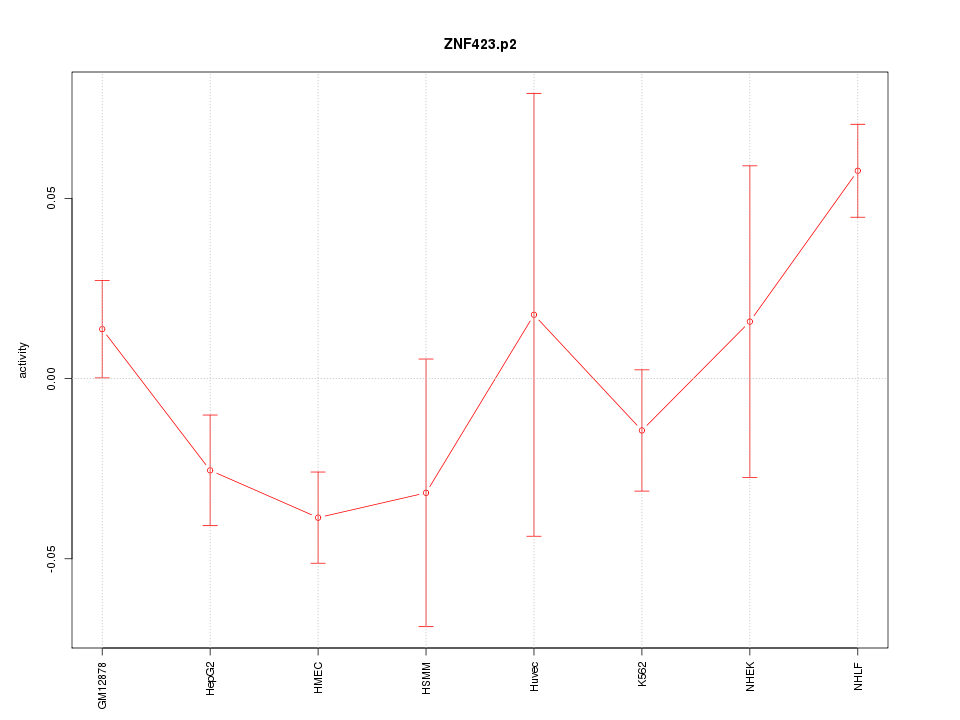
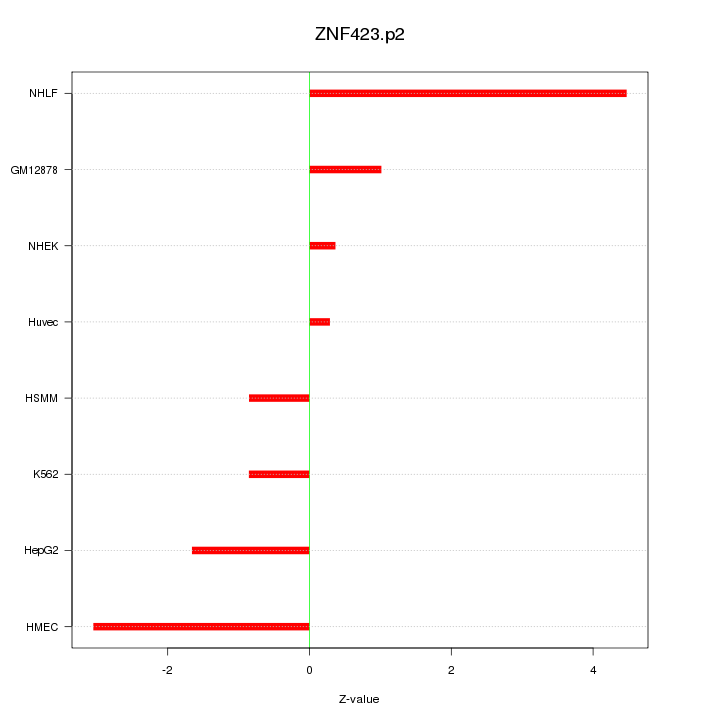
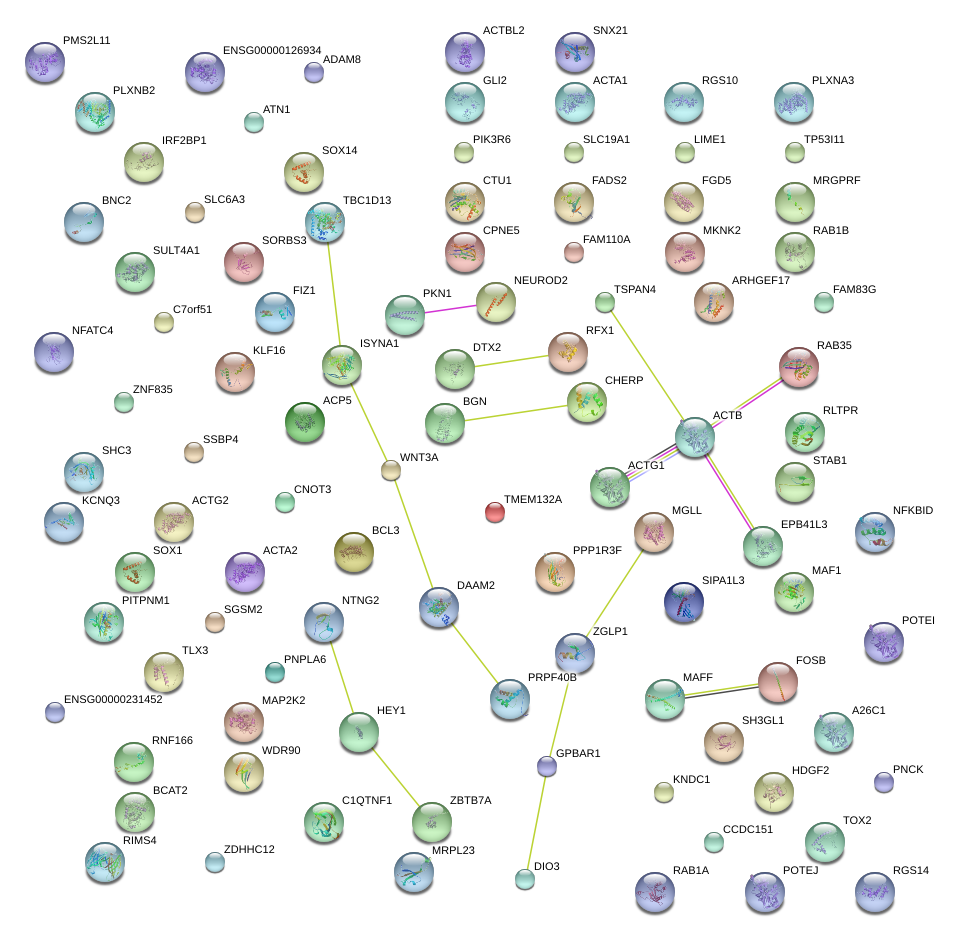
NEUROD2
|
neurogenic differentiation 2
|
|
chr19_+_14405100
|
1.765
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr19_-_4054200
|
1.709
|
|
MAP2K2
|
mitogen-activated protein kinase kinase 2
|
|
chr3_-_129024665
|
1.693
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr19_-_4017739
|
1.650
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr9_+_135880363
|
1.590
|
|
NCRNA00094
|
non-protein coding RNA 94
|
|
chr11_-_68537243
|
1.550
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr19_+_50663089
|
1.503
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr7_+_99919485
|
1.402
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr22_+_44828338
|
1.372
|
|
|
|
|
chr19_+_49943773
|
1.349
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr11_+_72696963
|
1.349
|
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr2_+_121271336
|
1.334
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr17_-_35017650
|
1.278
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr5_+_170668892
|
1.274
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr19_-_11406900
|
1.263
|
NM_145045
|
CCDC151
|
coiled-coil domain containing 151
|
|
chr19_-_11550765
|
1.263
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr17_+_74531873
|
1.235
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_+_18391423
|
1.205
|
|
|
|
|
chr9_-_130526192
|
1.201
|
NM_032799
|
ZDHHC12
|
zinc finger, DHHC-type containing 12
|
|
chr20_+_762355
|
1.194
|
NM_001042353
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr11_-_67028415
|
1.179
|
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr11_+_1925162
|
1.172
|
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr19_+_4423149
|
1.169
|
NM_001001520
NM_032631
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr11_+_65792603
|
1.168
|
NM_030981
|
RAB1B
|
RAB1B, member RAS oncogene family
|
|
chr2_+_100087812
|
1.166
|
|
|
|
|
chr10_-_134940315
|
1.140
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr19_+_7506632
|
1.126
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr19_-_1993022
|
1.122
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr5_+_176717449
|
1.118
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr19_-_1992862
|
1.106
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr14_+_23907065
|
1.102
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr9_+_134027154
|
1.090
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr17_+_2187545
|
1.066
|
NM_014853
NM_001098509
|
SGSM2
|
small G protein signaling modulator 2
|
|
chr11_-_44928803
|
1.048
|
NM_001076787
NM_006034
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr19_-_4351357
|
1.023
|
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr5_-_1498510
|
1.013
|
NM_001044
|
SLC6A3
|
solute carrier family 6 (neurotransmitter transporter, dopamine), member 3
|
|
chr11_+_834111
|
0.991
|
|
TSPAN4
|
tetraspanin 4
|
|
chr17_+_74531820
|
0.979
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr9_-_90983195
|
0.972
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr16_+_639278
|
0.971
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr11_+_72697310
|
0.964
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr5_+_86599575
|
0.961
|
|
|
|
|
chr12_-_119038941
|
0.954
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr16_-_3131348
|
0.952
|
|
|
|
|
chr19_+_43089720
|
0.949
|
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr8_-_80842452
|
0.947
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr11_+_60448487
|
0.946
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr16_+_66236512
|
0.936
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr19_-_18409971
|
0.931
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr19_-_54005916
|
0.925
|
NM_001164773
NM_001190
|
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial
|
|
chr19_-_10281220
|
0.921
|
NM_001103167
|
ZGLP1
|
zinc finger, GATA-like protein 1
|
|
chr2_+_218833982
|
0.920
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr19_+_43089681
|
0.919
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr19_+_18391211
|
0.916
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr11_+_61352209
|
0.916
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr12_+_48303607
|
0.902
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr19_+_14405216
|
0.902
|
|
PKN1
|
protein kinase N1
|
|
chr19_-_18409788
|
0.896
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr9_-_16860664
|
0.887
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_8711701
|
0.883
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr19_-_4351470
|
0.883
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr19_-_41083288
|
0.882
|
NM_139239
|
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chr12_-_119038995
|
0.880
|
NM_001167606
NM_006861
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr11_+_61352348
|
0.872
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr19_-_56303434
|
0.864
|
NM_145232
|
CTU1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe)
|
|
chr7_-_5536783
|
0.854
|
|
ACTB
|
actin, beta
|
|
chr20_+_61838479
|
0.854
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr1_-_226530350
|
0.851
|
|
|
|
|
chr8_+_22479115
|
0.846
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr10_+_134823925
|
0.845
|
NM_152643
|
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1
|
|
chr8_-_133561738
|
0.844
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr7_-_5536704
|
0.842
|
NM_001101
|
ACTB
|
actin, beta
|
|
chr6_+_39868126
|
0.835
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chrX_+_152413646
|
0.823
|
|
BGN
|
biglycan
|
|
chr8_+_145231386
|
0.816
|
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr14_+_101097440
|
0.816
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chrX_+_152413540
|
0.816
|
NM_001711
|
BGN
|
biglycan
|
|
chr20_+_61838389
|
0.813
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr19_-_1814546
|
0.809
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr1_+_226261374
|
0.806
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr3_+_14835472
|
0.804
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr18_-_5533967
|
0.803
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chrX_+_49013424
|
0.802
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr11_+_65792680
|
0.800
|
|
RAB1B
|
RAB1B, member RAS oncogene family
|
|
chr6_-_36915195
|
0.799
|
NM_020939
|
CPNE5
|
copine V
|
|
chr12_-_119038940
|
0.795
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr19_-_60802663
|
0.790
|
NM_032836
|
FIZ1
|
FLT3-interacting zinc finger 1
|
|
chr7_+_149733309
|
0.786
|
|
|
|
|
chr21_-_45786709
|
0.785
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr22_-_49062461
|
0.783
|
|
PLXNB2
|
plexin B2
|
|
chr12_-_119038836
|
0.779
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr16_-_87297525
|
0.775
|
NM_001171816
|
RNF166
|
ring finger protein 166
|
|
chr8_+_145231292
|
0.772
|
NM_032272
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr22_-_42589543
|
0.772
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chrX_+_153339814
|
0.771
|
NM_017514
|
PLXNA3
|
plexin A3
|
|
chr20_+_41978195
|
0.769
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr17_+_73738985
|
0.766
|
NM_001145529
|
TMEM235
|
transmembrane protein 235
|
|
chr7_+_75928907
|
0.757
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr9_+_130589330
|
0.744
|
NM_018201
|
TBC1D13
|
TBC1 domain family, member 13
|
|
chr17_-_18848713
|
0.743
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr3_+_52504410
|
0.736
|
|
STAB1
|
stabilin 1
|
|
chr8_+_121892719
|
0.734
|
|
|
|
|
chr19_-_61874934
|
0.731
|
|
ZNF835
|
zinc finger protein 835
|
|
chr19_-_1814425
|
0.730
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr19_-_13978096
|
0.730
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr20_+_43896178
|
0.727
|
|
SNX21
|
sorting nexin family member 21
|
|
chr12_+_6907646
|
0.723
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr22_+_36928972
|
0.723
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_+_59333232
|
0.722
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr19_-_16514232
|
0.722
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr20_-_42872295
|
0.716
|
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chrX_-_152592958
|
0.715
|
NM_001039582
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr8_-_133561933
|
0.708
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr10_-_121286034
|
0.708
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr3_+_138965844
|
0.705
|
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr19_-_51081147
|
0.705
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr11_+_1925075
|
0.702
|
NM_021134
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr16_+_66236559
|
0.696
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr19_+_40716153
|
0.695
|
NM_014364
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic
|
|
chr1_-_2333800
|
0.693
|
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr3_-_58538110
|
0.691
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr7_-_139987044
|
0.691
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr14_+_92049396
|
0.691
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr12_+_111829121
|
0.688
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr4_-_154929657
|
0.688
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr17_+_7252225
|
0.687
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr19_+_13111072
|
0.686
|
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr7_+_53070842
|
0.676
|
NM_182595
|
POM121L12
|
POM121 membrane glycoprotein-like 12
|
|
chr7_-_19151509
|
0.675
|
NM_152898
|
FERD3L
|
Fer3-like (Drosophila)
|
|
chr16_-_30453646
|
0.671
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr9_+_130589382
|
0.667
|
|
TBC1D13
|
TBC1 domain family, member 13
|
|
chr11_-_61415429
|
0.662
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr19_+_49866563
|
0.661
|
NM_001127893
NM_020219
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19
|
|
chr19_-_15172773
|
0.659
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr17_-_45582872
|
0.659
|
NM_032595
|
PPP1R9B
|
protein phosphatase 1, regulatory (inhibitor) subunit 9B
|
|
chr1_+_1437358
|
0.659
|
NM_001170535
NM_018188
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr1_-_16272523
|
0.658
|
NM_182623
|
FAM131C
|
family with sequence similarity 131, member C
|
|
chr22_+_35639564
|
0.654
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr16_-_87300233
|
0.651
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr9_+_95378492
|
0.650
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr1_-_9052253
|
0.649
|
NM_001135585
NM_003039
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr12_+_109956210
|
0.647
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr15_+_38973893
|
0.646
|
NM_020857
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae)
|
|
chr19_-_16514202
|
0.643
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr9_-_16860719
|
0.643
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr1_+_99502655
|
0.642
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr19_-_16514256
|
0.641
|
NM_006387
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr3_-_16530132
|
0.639
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr8_+_120497732
|
0.639
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr3_+_52504391
|
0.634
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr10_+_99069007
|
0.633
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr6_+_33352886
|
0.632
|
NM_003782
|
B3GALT4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4
|
|
chr7_-_1510533
|
0.628
|
NM_001080453
|
INTS1
|
integrator complex subunit 1
|
|
chr8_-_70908130
|
0.627
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr19_-_16514247
|
0.627
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr16_-_1783653
|
0.626
|
NM_001146006
NM_004970
|
IGFALS
|
insulin-like growth factor binding protein, acid labile subunit
|
|
chr19_-_52426055
|
0.626
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr1_-_27825178
|
0.625
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr1_-_44593526
|
0.624
|
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr19_-_5573794
|
0.621
|
NM_014649
|
SAFB2
|
scaffold attachment factor B2
|
|
chr5_+_175241449
|
0.620
|
|
CPLX2
|
complexin 2
|
|
chr7_-_150129295
|
0.616
|
NM_014020
|
TMEM176B
|
transmembrane protein 176B
|
|
chr19_+_53589943
|
0.616
|
NM_000836
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D
|
|
chr6_-_109868400
|
0.615
|
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr12_-_131415854
|
0.613
|
NM_001122636
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr1_-_2333863
|
0.612
|
NM_002617
NM_153818
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr16_-_87300308
|
0.610
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr19_-_44214972
|
0.608
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr8_-_65873658
|
0.607
|
NM_004820
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1
|
|
chr3_+_13496714
|
0.603
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr22_+_35586950
|
0.601
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr17_+_7549138
|
0.600
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr16_+_53915971
|
0.599
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chrX_+_70899864
|
0.597
|
NM_001145140
NM_001145139
|
CXorf49
CXorf49B
|
chromosome X open reading frame 49
chromosome X open reading frame 49B
|
|
chr19_+_14108974
|
0.597
|
|
|
|
|
chrX_+_152608159
|
0.595
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr8_+_145137354
|
0.592
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr10_-_120345148
|
0.591
|
NM_004248
|
PRLHR
|
prolactin releasing hormone receptor
|
|
chr20_-_43972982
|
0.590
|
|
PLTP
|
phospholipid transfer protein
|
|
chr1_+_1437400
|
0.590
|
|
ATAD3A
ATAD3B
|
ATPase family, AAA domain containing 3A
ATPase family, AAA domain containing 3B
|
|
chr19_+_43502360
|
0.589
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_-_1518875
|
0.585
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr20_+_29526751
|
0.585
|
NM_014012
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1
|
|
chr19_-_5291700
|
0.584
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr3_-_129023850
|
0.581
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr22_+_48740432
|
0.580
|
|
PIM3
|
pim-3 oncogene
|
|
chr12_+_131577229
|
0.577
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr8_+_144711619
|
0.574
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr16_-_30453499
|
0.572
|
|
ZNF747
|
zinc finger protein 747
|
|
chr14_-_67051773
|
0.570
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr19_+_52940604
|
0.569
|
NM_015710
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr8_+_142207866
|
0.569
|
NM_014957
|
DENND3
|
DENN/MADD domain containing 3
|
|
chr10_+_3099708
|
0.568
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr8_-_22022757
|
0.555
|
NM_024815
|
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18
|
|
chr3_-_16529996
|
0.551
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr5_+_131774478
|
0.551
|
NM_001013717
|
C5orf56
|
chromosome 5 open reading frame 56
|
|
chrX_+_136475979
|
0.550
|
NM_003413
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr5_+_68823874
|
0.550
|
NM_002538
|
OCLN
|
occludin
|
|
chr7_-_1144347
|
0.546
|
NM_001134395
NM_001134396
NM_032350
|
C7orf50
|
chromosome 7 open reading frame 50
|