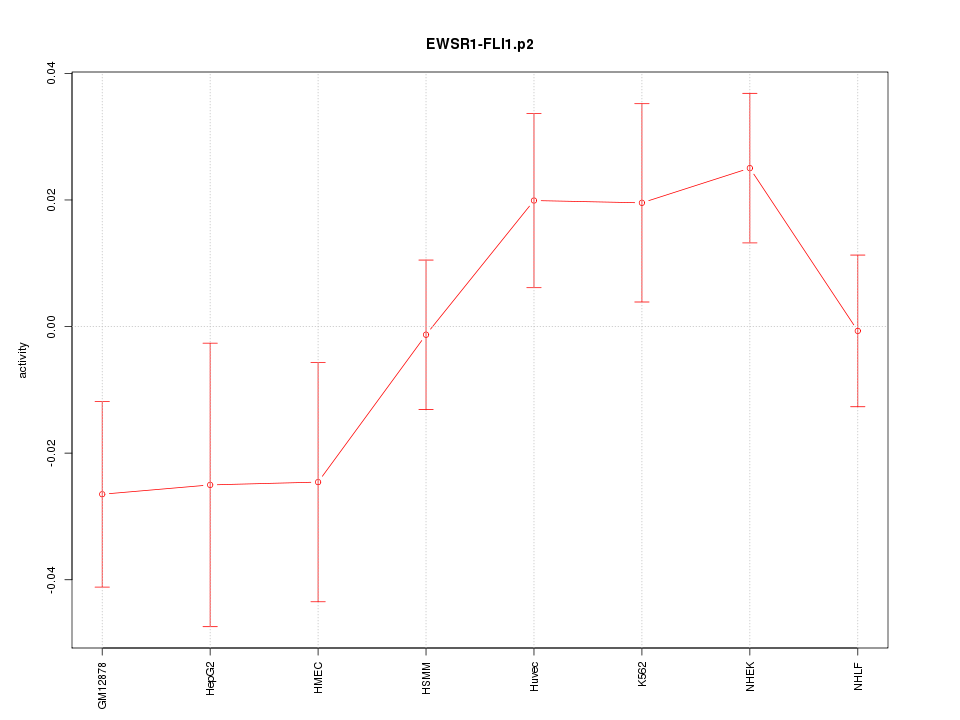
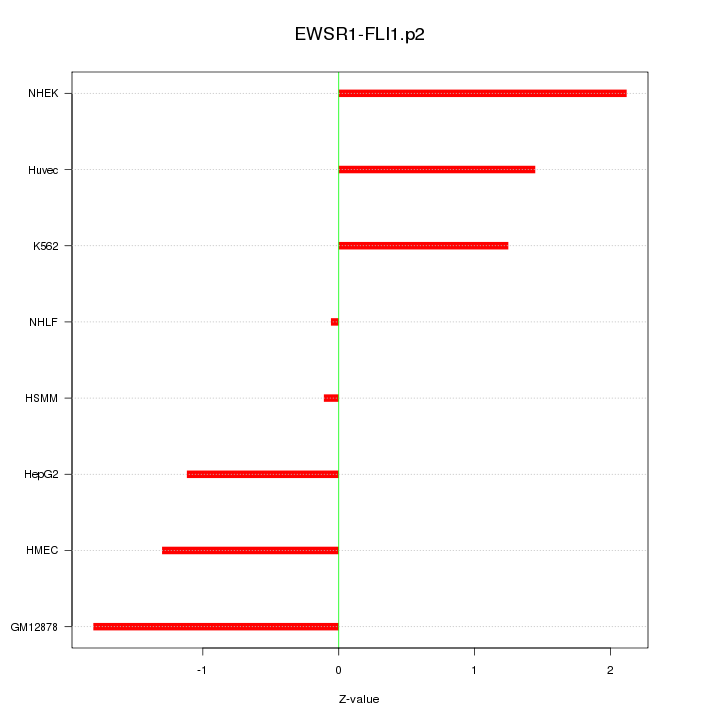
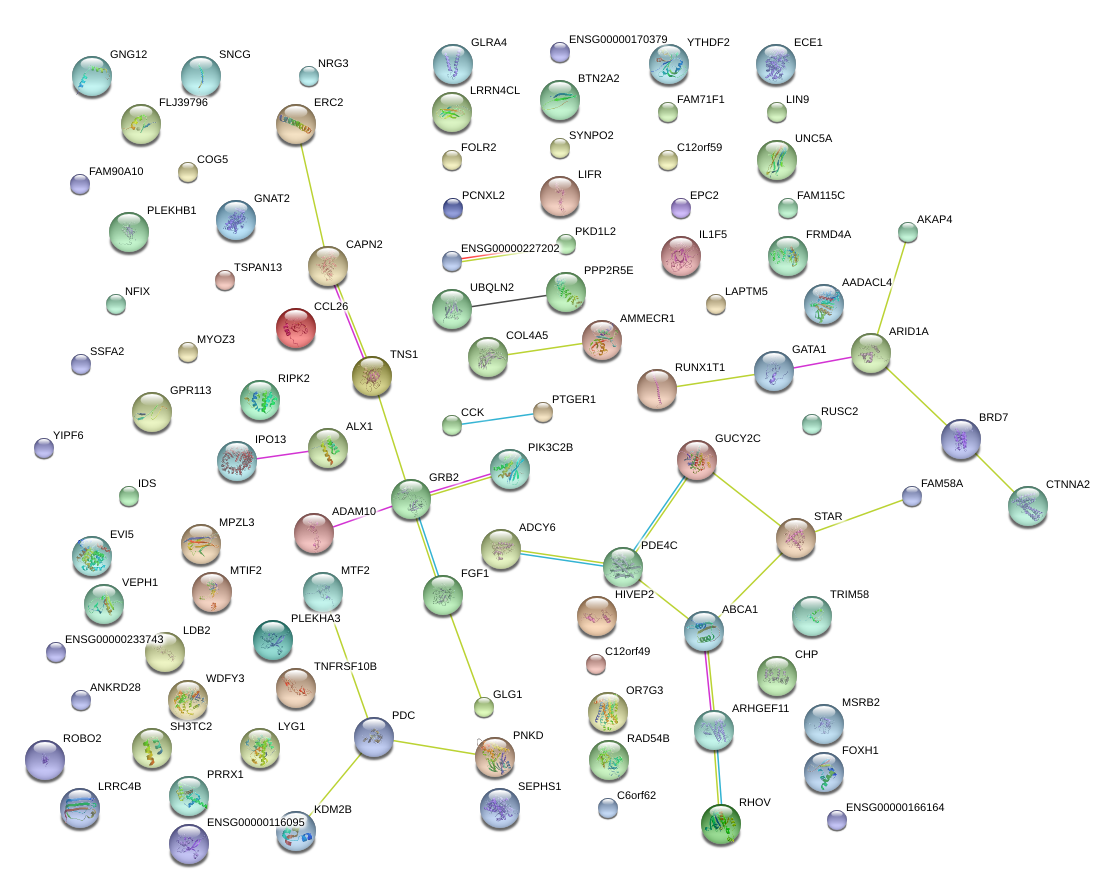
|
chr1_-_93030538
|
5.359
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr1_+_221955876
|
3.208
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr5_-_148422799
|
2.171
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr19_-_9098625
|
1.498
|
NM_001001958
|
OR7G3
|
olfactory receptor, family 7, subfamily G, member 3
|
|
chr19_-_55763091
|
1.378
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr11_+_73034870
|
1.341
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr10_+_23424383
|
1.293
|
NM_012228
|
MSRB2
|
methionine sulfoxide reductase B2
|
|
chr1_-_155281785
|
1.149
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chrX_-_148394570
|
1.148
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr20_-_48741481
|
1.132
|
|
|
|
|
chr1_+_12627152
|
1.120
|
NM_001013630
|
AADACL4
|
arylacetamide deacetylase-like 4
|
|
chr9_+_35528890
|
1.091
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_143043871
|
1.000
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr11_-_62213946
|
0.998
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_68071635
|
0.991
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr6_-_143307976
|
0.991
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr12_-_115660017
|
0.928
|
NM_024738
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr12_+_88627601
|
0.878
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr8_-_93176819
|
0.864
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_49851743
|
0.841
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr4_+_120029426
|
0.836
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr2_+_113532685
|
0.804
|
NM_173170
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr8_-_93177057
|
0.793
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_15875932
|
0.785
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_31698447
|
0.746
|
|
|
|
|
chr16_-_48960186
|
0.709
|
NM_001173984
NM_013263
|
BRD7
|
bromodomain containing 7
|
|
chr2_-_26395420
|
0.703
|
NM_001145168
NM_001145169
|
GPR113
|
G protein-coupled receptor 113
|
|
chr10_+_83627422
|
0.701
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr1_-_224564038
|
0.685
|
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chr2_+_113533155
|
0.661
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr8_-_95556460
|
0.646
|
NM_012415
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr9_+_35528628
|
0.628
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_+_79593567
|
0.600
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr2_-_55349851
|
0.570
|
NM_002453
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr6_-_24827265
|
0.568
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr16_-_73198416
|
0.562
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr7_+_21434818
|
0.540
|
|
|
|
|
chr5_-_38631263
|
0.539
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr2_+_179053512
|
0.529
|
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3
|
|
chrX_-_109447970
|
0.525
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr3_-_42282665
|
0.519
|
NM_000729
|
CCK
|
cholecystokinin
|
|
chr19_+_12996830
|
0.518
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr5_-_141981083
|
0.517
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chrX_+_107569671
|
0.517
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr1_-_109957195
|
0.512
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chrX_+_48529905
|
0.508
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr15_-_56829225
|
0.493
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr10_+_88708267
|
0.490
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr7_-_106991941
|
0.479
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chrX_-_109448083
|
0.478
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr5_-_64100239
|
0.477
|
NM_173829
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr8_-_38127489
|
0.473
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr7_+_128142661
|
0.472
|
NM_032599
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chrX_-_109447954
|
0.458
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr8_-_145671947
|
0.446
|
|
FOXH1
|
forkhead box H1
|
|
chr1_+_246087104
|
0.443
|
NM_015431
|
TRIM58
|
tripartite motif containing 58
|
|
chr12_+_10222823
|
0.441
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr7_-_75256999
|
0.436
|
NM_006072
|
CCL26
|
chemokine (C-C motif) ligand 26
|
|
chr11_-_117628211
|
0.428
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr1_-_31003181
|
0.420
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr2_+_149118791
|
0.416
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_-_99284070
|
0.415
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr12_-_47464143
|
0.414
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr5_-_71838875
|
0.411
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr16_-_73198498
|
0.407
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr1_-_21478571
|
0.406
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_-_218551786
|
0.403
|
|
TNS1
|
tensin 1
|
|
chrX_-_102870207
|
0.400
|
NM_001024452
NM_001172285
|
GLRA4
|
glycine receptor, alpha 4
|
|
chr1_-_202730135
|
0.399
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr7_+_16759845
|
0.398
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr16_-_73198515
|
0.398
|
NM_001145666
NM_001145667
NM_012201
|
GLG1
|
golgi glycoprotein 1
|
|
chr16_-_79811475
|
0.394
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr1_+_246087230
|
0.393
|
|
TRIM58
|
tripartite motif containing 58
|
|
chr11_+_71605466
|
0.392
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr9_-_106705867
|
0.390
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_+_182464910
|
0.389
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr3_-_158704056
|
0.385
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr4_-_86106567
|
0.381
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_-_231498081
|
0.381
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr12_+_84198015
|
0.381
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr17_-_70901303
|
0.379
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr3_+_77171938
|
0.375
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr14_-_63079299
|
0.372
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr5_-_64100176
|
0.371
|
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr19_-_18220009
|
0.370
|
NM_000923
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr10_-_13429397
|
0.363
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr8_-_22982435
|
0.363
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr6_+_26491302
|
0.361
|
NM_001197238
NM_001197239
NM_001197240
NM_006995
NM_181531
NM_001197237
|
BTN2A2
|
butyrophilin, subfamily 2, member A2
|
|
chr1_-_184684478
|
0.358
|
NM_022576
|
PDC
|
phosducin
|
|
chr8_-_145672525
|
0.358
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr19_+_12996745
|
0.354
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_120503241
|
0.351
|
NM_032590
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr1_+_26895108
|
0.350
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr1_-_224563820
|
0.344
|
NM_173083
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chr2_-_55349818
|
0.344
|
NM_001005369
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr3_-_56477404
|
0.342
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chrX_+_56606686
|
0.338
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr1_+_93317374
|
0.337
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr19_-_14447122
|
0.334
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr2_+_182464713
|
0.329
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr1_+_44185005
|
0.326
|
NM_014652
|
IPO13
|
importin 13
|
|
chr11_-_62213577
|
0.322
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_224563461
|
0.320
|
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chrX_+_67635348
|
0.316
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr1_+_168898898
|
0.309
|
|
PRRX1
|
paired related homeobox 1
|
|
chr15_+_39310757
|
0.306
|
|
CHP
|
calcium binding protein P22
|
|
chr8_-_93176618
|
0.296
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_-_14412813
|
0.296
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr15_-_56829069
|
0.290
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr4_-_16509354
|
0.279
|
|
LDB2
|
LIM domain binding 2
|
|
chr1_+_28935719
|
0.278
|
NM_001173128
NM_016258
|
YTHDF2
|
YTH domain family, member 2
|
|
chr5_+_150020595
|
0.278
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr9_-_106705780
|
0.275
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_+_33807168
|
0.273
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chrX_-_107569256
|
0.271
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr7_-_99365178
|
0.269
|
NM_181538
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa
|
|
chr21_+_44544392
|
0.268
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr11_+_2279932
|
0.263
|
|
TSPAN32
|
tetraspanin 32
|
|
chr3_-_188936941
|
0.263
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_+_54621499
|
0.261
|
NM_001195256
|
LOC645971
|
hypothetical protein LOC645971
|
|
chr20_+_41621079
|
0.260
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr10_-_103864651
|
0.260
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr15_-_66285427
|
0.255
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr4_-_16509446
|
0.254
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr10_+_104211134
|
0.253
|
|
TMEM180
|
transmembrane protein 180
|
|
chr11_-_74119833
|
0.251
|
NM_015424
|
CHRDL2
|
chordin-like 2
|
|
chr19_-_8581559
|
0.249
|
NM_030957
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10
|
|
chr11_+_58668827
|
0.246
|
NM_022074
NM_198847
|
FAM111A
|
family with sequence similarity 111, member A
|
|
chr9_-_129679760
|
0.245
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr15_+_81907176
|
0.243
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr15_+_81906983
|
0.242
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr10_+_70553862
|
0.237
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_+_28936030
|
0.237
|
NM_001172828
|
YTHDF2
|
YTH domain family, member 2
|
|
chr19_-_242168
|
0.236
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr7_-_94123308
|
0.235
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr11_+_10434056
|
0.233
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr2_-_128155829
|
0.233
|
NM_001161404
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr18_+_54682261
|
0.230
|
|
ZNF532
|
zinc finger protein 532
|
|
chr16_-_69277194
|
0.226
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr5_+_35892747
|
0.223
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr18_+_75388522
|
0.218
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr9_+_134843918
|
0.218
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr7_+_36395913
|
0.217
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr9_-_125732005
|
0.214
|
NM_020946
NM_024820
|
DENND1A
|
DENN/MADD domain containing 1A
|
|
chr10_+_104211148
|
0.214
|
NM_024789
|
TMEM180
|
transmembrane protein 180
|
|
chr19_-_44518465
|
0.213
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr15_+_89248423
|
0.213
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr15_+_72861767
|
0.211
|
|
CSK
|
c-src tyrosine kinase
|
|
chr11_-_74120077
|
0.210
|
|
CHRDL2
|
chordin-like 2
|
|
chr12_+_54947293
|
0.206
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr21_+_44544381
|
0.206
|
|
PFKL
|
phosphofructokinase, liver
|
|
chrX_-_131918905
|
0.205
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr10_+_75211810
|
0.204
|
NM_203298
|
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1
|
|
chr11_-_76800525
|
0.201
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr1_-_31003260
|
0.199
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chrX_+_24077649
|
0.197
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chrX_+_152252738
|
0.196
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr10_-_35143900
|
0.196
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr3_+_143925904
|
0.195
|
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr11_-_65244486
|
0.195
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr4_-_16509126
|
0.193
|
|
LDB2
|
LIM domain binding 2
|
|
chr19_-_55914837
|
0.192
|
|
|
|
|
chr7_+_8440109
|
0.191
|
NM_152745
|
NXPH1
|
neurexophilin 1
|
|
chr13_+_28131279
|
0.188
|
|
POMP
|
proteasome maturation protein
|
|
chrX_-_85189194
|
0.183
|
NM_000390
NM_001145414
|
CHM
|
choroideremia (Rab escort protein 1)
|
|
chr3_-_120878910
|
0.182
|
NM_005694
|
COX17
|
COX17 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr3_+_188131209
|
0.180
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr17_-_8004671
|
0.179
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr19_-_14447163
|
0.178
|
NM_000955
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr11_+_68614059
|
0.177
|
|
|
|
|
chrX_-_133757901
|
0.176
|
NM_001170756
NM_145284
|
FAM122B
|
family with sequence similarity 122B
|
|
chr13_+_28131140
|
0.169
|
NM_015932
|
POMP
|
proteasome maturation protein
|
|
chr19_+_54883732
|
0.167
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr16_-_45565068
|
0.165
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr11_+_2279812
|
0.165
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr15_+_39310693
|
0.165
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr3_+_171558215
|
0.164
|
|
SKIL
|
SKI-like oncogene
|
|
chr6_-_32251877
|
0.164
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr21_+_44544349
|
0.160
|
NM_002626
|
PFKL
|
phosphofructokinase, liver
|
|
chr16_-_67723511
|
0.160
|
NM_001040146
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr12_+_92295738
|
0.156
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr12_+_44409762
|
0.155
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr2_-_104834344
|
0.154
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr3_+_33130563
|
0.152
|
|
CRTAP
|
cartilage associated protein
|
|
chr12_+_54947180
|
0.152
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr2_+_79593679
|
0.151
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr10_+_70553936
|
0.149
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrX_+_133758108
|
0.149
|
NM_001170780
|
FAM122C
|
family with sequence similarity 122C
|
|
chr3_+_188130967
|
0.148
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr11_+_10433241
|
0.148
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr6_-_130578110
|
0.148
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr20_+_41621119
|
0.147
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr3_+_33130451
|
0.147
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr14_-_99016833
|
0.147
|
|
SETD3
|
SET domain containing 3
|
|
chr20_+_10147455
|
0.146
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr10_-_103864579
|
0.145
|
|
LDB1
|
LIM domain binding 1
|
|
chr12_+_44409743
|
0.144
|
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr5_+_137701601
|
0.141
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr12_-_91346054
|
0.141
|
NM_001025232
|
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand
|
|
chr19_+_60487345
|
0.140
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|