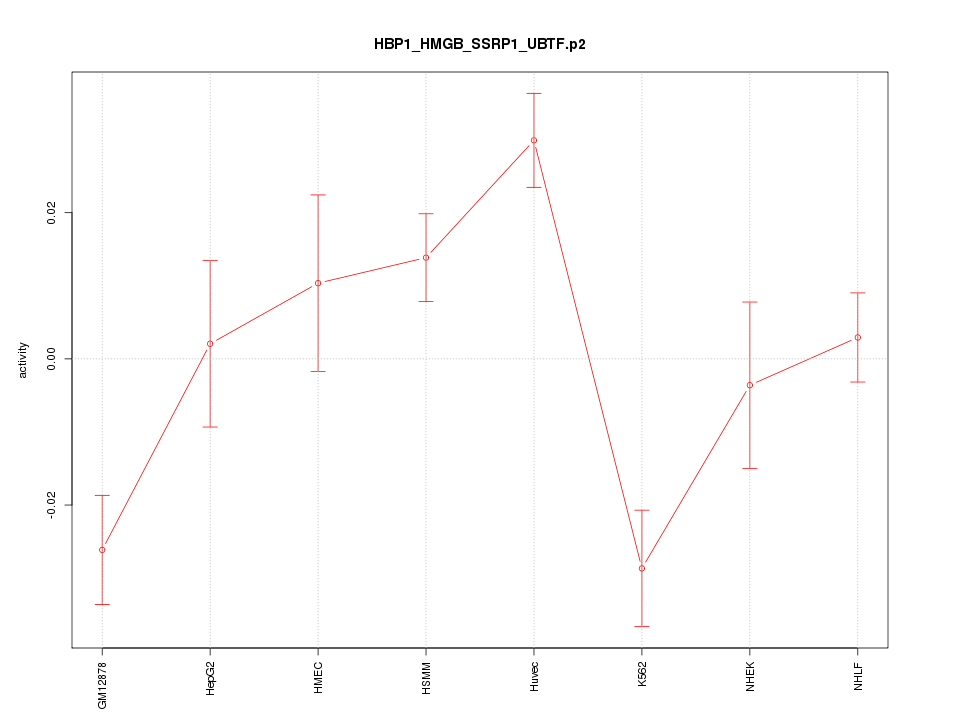
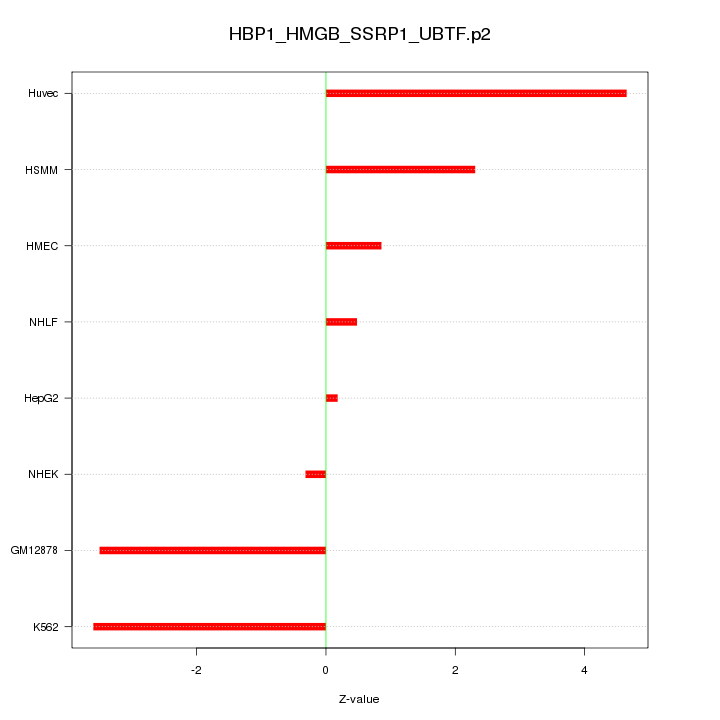
|
chr11_+_129496792
|
7.019
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr3_-_116272911
|
5.643
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_111995213
|
5.090
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr4_-_101658094
|
4.958
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chrX_-_19598912
|
4.685
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_-_27186400
|
4.484
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr4_+_141397889
|
4.318
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr2_-_162808123
|
3.991
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr16_+_85169615
|
3.926
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr3_-_150533948
|
3.857
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr1_-_177106702
|
3.793
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr11_-_85108030
|
3.727
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr2_-_56266408
|
3.688
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr9_-_23816062
|
3.661
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr18_+_7936885
|
3.493
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr8_+_70567634
|
3.302
|
|
SULF1
|
sulfatase 1
|
|
chr9_+_18464090
|
3.298
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr8_+_70567581
|
3.277
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr8_-_93144039
|
3.242
|
|
|
|
|
chr11_+_57547928
|
3.239
|
NM_001005212
|
OR9Q1
|
olfactory receptor, family 9, subfamily Q, member 1
|
|
chr3_-_11585264
|
3.169
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr12_-_16649211
|
3.139
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_+_102380156
|
3.135
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr2_-_72228427
|
3.032
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr7_+_134114912
|
3.000
|
|
CALD1
|
caldesmon 1
|
|
chr9_+_18464138
|
2.986
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr8_-_93144366
|
2.956
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_81544432
|
2.942
|
|
LPHN2
|
latrophilin 2
|
|
chr7_-_83662152
|
2.861
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr20_+_19818209
|
2.837
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr3_+_174598937
|
2.796
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr5_-_111119846
|
2.768
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr15_+_37330176
|
2.760
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chr11_-_85107569
|
2.750
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_+_676640
|
2.750
|
|
|
|
|
chr7_-_27153892
|
2.748
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr6_+_21774624
|
2.738
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr3_+_148593911
|
2.721
|
|
|
|
|
chr3_-_188936941
|
2.717
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr7_+_134114957
|
2.676
|
|
CALD1
|
caldesmon 1
|
|
chr8_-_108579270
|
2.675
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_83052606
|
2.663
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr10_-_118887801
|
2.644
|
NM_001112704
NM_199131
|
VAX1
|
ventral anterior homeobox 1
|
|
chr6_+_153060722
|
2.629
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr14_+_36196523
|
2.625
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr3_+_173043832
|
2.569
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr7_-_27108826
|
2.512
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr2_-_128138435
|
2.510
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr6_+_148705421
|
2.503
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr10_+_104525877
|
2.491
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr1_-_242073094
|
2.468
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr16_+_65938753
|
2.445
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chrX_+_135079461
|
2.409
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_-_56824672
|
2.403
|
|
DST
|
dystonin
|
|
chr12_-_10142779
|
2.355
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr3_+_182900081
|
2.332
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr2_+_108571336
|
2.317
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_38631263
|
2.294
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr15_+_45797977
|
2.294
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr7_-_27149750
|
2.290
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr13_-_32678142
|
2.276
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr9_+_27099138
|
2.248
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr16_+_1068781
|
2.244
|
NM_001053
|
SSTR5
|
somatostatin receptor 5
|
|
chr14_+_76298693
|
2.233
|
|
VASH1
|
vasohibin 1
|
|
chr15_-_53996597
|
2.213
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr21_-_27261276
|
2.186
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr8_-_6407882
|
2.165
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr10_-_116434364
|
2.159
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_-_115825742
|
2.148
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_44424423
|
2.135
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chrX_+_15428820
|
2.135
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr6_-_94185780
|
2.133
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr6_-_134540666
|
2.127
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr8_-_18710575
|
2.125
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr20_-_45418818
|
2.123
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr13_+_23742846
|
2.114
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr7_-_11838260
|
2.112
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr5_+_170668892
|
2.108
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr17_+_72795567
|
2.044
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr6_-_139654807
|
2.013
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr6_-_56824371
|
2.008
|
|
DST
|
dystonin
|
|
chr1_-_152431175
|
2.007
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr15_-_68781662
|
2.000
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr6_+_31190505
|
1.965
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr9_-_16717887
|
1.952
|
|
BNC2
|
basonuclin 2
|
|
chr13_-_35327798
|
1.943
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr18_+_71051713
|
1.935
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr18_-_29412148
|
1.931
|
|
|
|
|
chr5_-_39400319
|
1.911
|
NM_001737
|
C9
|
complement component 9
|
|
chr3_+_152970904
|
1.881
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr17_+_72380323
|
1.869
|
NM_198955
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr17_-_46479127
|
1.856
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr12_-_6103945
|
1.846
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chrX_-_33267646
|
1.841
|
NM_000109
|
DMD
|
dystrophin
|
|
chr5_-_158567411
|
1.839
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr1_+_22107137
|
1.834
|
|
RPL21
|
ribosomal protein L21
|
|
chr3_-_69254226
|
1.834
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr14_-_88953059
|
1.823
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr14_-_91483325
|
1.822
|
|
FBLN5
|
fibulin 5
|
|
chr10_+_123913094
|
1.810
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr8_-_93099040
|
1.808
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_82557956
|
1.800
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_+_102340579
|
1.795
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr13_+_99431417
|
1.779
|
|
|
|
|
chr6_-_138935359
|
1.778
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chrX_+_135079120
|
1.773
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_+_15145001
|
1.769
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_+_112337199
|
1.766
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr9_-_15240113
|
1.762
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr12_+_32546243
|
1.752
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr2_+_33213111
|
1.738
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_-_50573759
|
1.727
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr5_-_125956996
|
1.726
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr4_-_186814852
|
1.713
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_-_8723893
|
1.712
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr7_-_27125738
|
1.696
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr3_-_71376919
|
1.685
|
|
FOXP1
|
forkhead box P1
|
|
chr21_-_31853160
|
1.677
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr20_+_11846536
|
1.666
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr17_-_1359745
|
1.658
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr4_-_20594645
|
1.657
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr7_+_134114701
|
1.655
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr17_-_71017509
|
1.650
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr8_+_40130143
|
1.649
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr9_+_117955890
|
1.647
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_+_13240983
|
1.642
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr5_+_15553288
|
1.637
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr20_+_31613800
|
1.609
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr13_-_98428244
|
1.607
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_+_45894480
|
1.588
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr12_+_83954229
|
1.586
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chrX_+_152684150
|
1.585
|
|
PLXNB3
|
plexin B3
|
|
chr12_-_44644216
|
1.584
|
|
|
|
|
chrX_+_77889861
|
1.572
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr4_-_73653331
|
1.572
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr12_+_13240987
|
1.563
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr17_+_72376022
|
1.549
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr12_-_120725210
|
1.545
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr10_+_80672842
|
1.539
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr4_+_146820805
|
1.538
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr18_+_29412538
|
1.538
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr2_-_213111525
|
1.529
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr5_-_16795336
|
1.516
|
|
MYO10
|
myosin X
|
|
chr2_-_73170290
|
1.507
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr7_-_41709191
|
1.506
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr21_-_38792173
|
1.492
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr2_-_183095308
|
1.490
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr4_-_152366970
|
1.487
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr13_-_32658215
|
1.482
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_-_147266257
|
1.473
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr6_-_56615544
|
1.462
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr1_+_176577228
|
1.459
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr9_-_4289915
|
1.457
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr5_-_83715947
|
1.437
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr7_+_134226690
|
1.437
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chrX_-_62891066
|
1.425
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr1_+_161305188
|
1.425
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_36777336
|
1.423
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr10_-_32707549
|
1.414
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr6_-_42218692
|
1.412
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr14_+_57964462
|
1.409
|
NM_014749
|
KIAA0586
|
KIAA0586
|
|
chr10_-_103864579
|
1.408
|
|
LDB1
|
LIM domain binding 1
|
|
chr2_-_133144094
|
1.403
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr12_-_83954185
|
1.401
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr4_-_152368492
|
1.399
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_+_119174947
|
1.392
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr5_+_149960820
|
1.391
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr9_-_129557373
|
1.390
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr1_-_68470585
|
1.388
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr11_+_20001130
|
1.386
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr5_-_83716346
|
1.386
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr9_+_35896188
|
1.385
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr4_+_87070449
|
1.383
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_+_130719034
|
1.381
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr2_-_213723298
|
1.370
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr14_+_62740878
|
1.367
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr7_+_134226727
|
1.354
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_175896829
|
1.352
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr2_-_188021117
|
1.349
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr1_+_226404037
|
1.347
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr8_+_32623792
|
1.345
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr10_+_101532450
|
1.341
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr9_-_72673753
|
1.333
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chrX_-_31436335
|
1.328
|
NM_004014
|
DMD
|
dystrophin
|
|
chr16_+_54072969
|
1.319
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr10_-_123346148
|
1.311
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr6_-_74284522
|
1.307
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr3_-_49372584
|
1.306
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr9_-_116190038
|
1.302
|
|
AKNA
|
AT-hook transcription factor
|
|
chr2_-_134042500
|
1.302
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr17_-_36346240
|
1.302
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_+_20000938
|
1.301
|
|
NAV2
|
neuron navigator 2
|
|
chr2_-_133145539
|
1.293
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr11_-_121492010
|
1.288
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr9_+_124835608
|
1.283
|
|
RABGAP1
GPR21
|
RAB GTPase activating protein 1
G protein-coupled receptor 21
|
|
chr4_+_160408447
|
1.281
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr3_-_48576204
|
1.268
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr4_-_138673078
|
1.267
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr11_+_101488380
|
1.263
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr6_-_89984214
|
1.256
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|