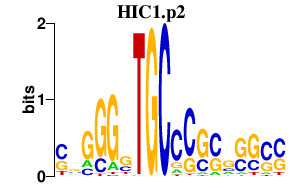
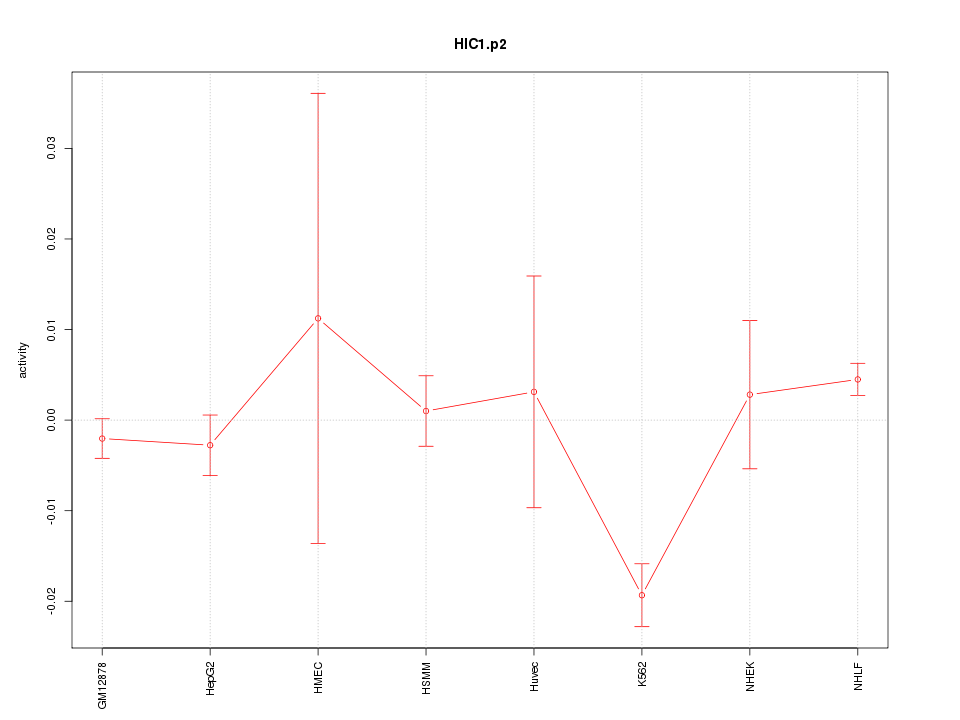
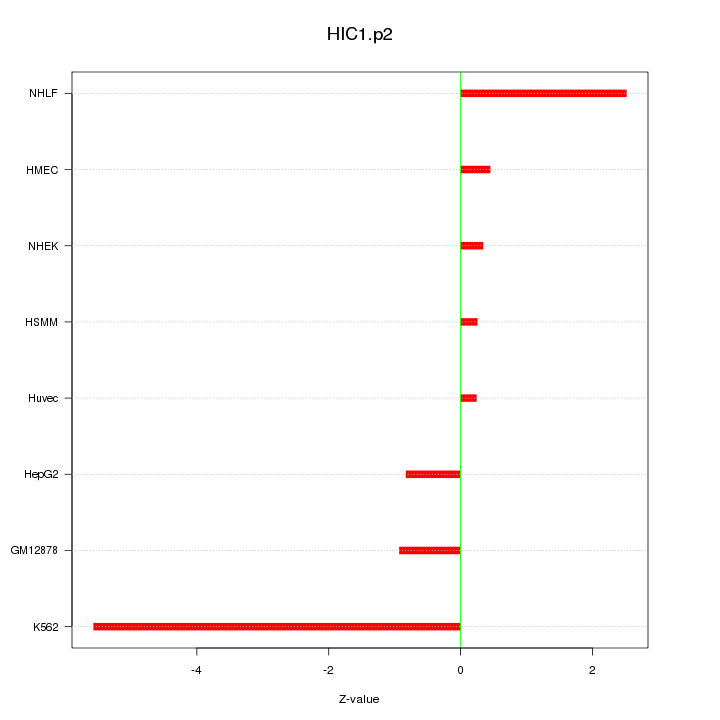
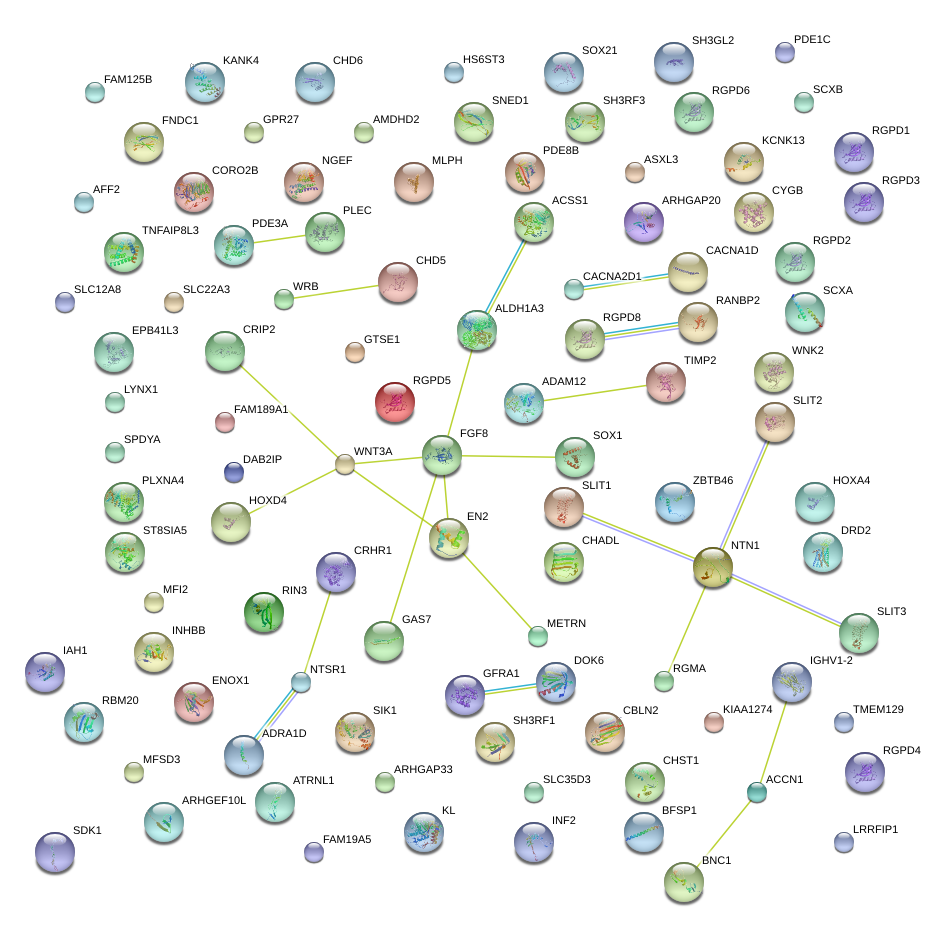
|
1.54
|
8.28e-29
|
GO:0007399
|
nervous system development
|
|
1.30
|
9.48e-28
|
GO:0032502
|
developmental process
|
|
1.32
|
1.84e-26
|
GO:0007275
|
multicellular organismal development
|
|
1.42
|
1.36e-25
|
GO:0030154
|
cell differentiation
|
|
1.41
|
5.75e-25
|
GO:0048869
|
cellular developmental process
|
|
1.64
|
5.29e-24
|
GO:0022008
|
neurogenesis
|
|
1.48
|
3.28e-23
|
GO:0009653
|
anatomical structure morphogenesis
|
|
1.64
|
2.30e-22
|
GO:0048699
|
generation of neurons
|
|
1.33
|
2.56e-22
|
GO:0048731
|
system development
|
|
1.30
|
7.52e-22
|
GO:0048856
|
anatomical structure development
|
|
1.59
|
3.09e-20
|
GO:0048468
|
cell development
|
|
1.80
|
3.65e-19
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
|
1.66
|
7.90e-19
|
GO:0030182
|
neuron differentiation
|
|
1.71
|
1.38e-17
|
GO:0048666
|
neuron development
|
|
1.81
|
4.84e-17
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
|
1.82
|
1.07e-16
|
GO:0007409
|
axonogenesis
|
|
1.68
|
1.15e-16
|
GO:0000902
|
cell morphogenesis
|
|
1.79
|
2.22e-16
|
GO:0048812
|
neuron projection morphogenesis
|
|
1.65
|
4.11e-16
|
GO:0032989
|
cellular component morphogenesis
|
|
1.74
|
7.31e-16
|
GO:0031175
|
neuron projection development
|
|
1.72
|
1.33e-15
|
GO:0032990
|
cell part morphogenesis
|
|
1.72
|
2.19e-15
|
GO:0048858
|
cell projection morphogenesis
|
|
1.09
|
3.66e-15
|
GO:0009987
|
cellular process
|
|
1.62
|
8.20e-15
|
GO:0030030
|
cell projection organization
|
|
1.14
|
4.95e-14
|
GO:0050794
|
regulation of cellular process
|
|
1.36
|
1.95e-13
|
GO:0023051
|
regulation of signaling
|
|
1.13
|
2.76e-13
|
GO:0065007
|
biological regulation
|
|
1.13
|
4.23e-12
|
GO:0050789
|
regulation of biological process
|
|
1.79
|
4.98e-12
|
GO:0007411
|
axon guidance
|
|
1.58
|
1.11e-11
|
GO:0009887
|
organ morphogenesis
|
|
1.63
|
1.58e-11
|
GO:0007417
|
central nervous system development
|
|
1.30
|
6.58e-11
|
GO:0048513
|
organ development
|
|
1.75
|
3.60e-10
|
GO:0001501
|
skeletal system development
|
|
1.39
|
1.32e-09
|
GO:0010646
|
regulation of cell communication
|
|
1.73
|
1.56e-09
|
GO:0060284
|
regulation of cell development
|
|
1.33
|
4.20e-09
|
GO:0009966
|
regulation of signal transduction
|
|
1.67
|
4.71e-09
|
GO:0007420
|
brain development
|
|
1.48
|
5.05e-09
|
GO:0045595
|
regulation of cell differentiation
|
|
1.48
|
5.98e-09
|
GO:0009790
|
embryo development
|
|
1.39
|
9.99e-09
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
|
1.73
|
1.11e-08
|
GO:0048729
|
tissue morphogenesis
|
|
1.75
|
1.23e-08
|
GO:0035295
|
tube development
|
|
1.65
|
1.53e-08
|
GO:0048598
|
embryonic morphogenesis
|
|
1.18
|
2.32e-08
|
GO:0023052
|
signaling
|
|
1.78
|
4.44e-08
|
GO:0050767
|
regulation of neurogenesis
|
|
1.72
|
7.31e-08
|
GO:0051960
|
regulation of nervous system development
|
|
1.39
|
9.12e-08
|
GO:0050793
|
regulation of developmental process
|
|
1.43
|
1.55e-07
|
GO:2000026
|
regulation of multicellular organismal development
|
|
1.14
|
2.56e-07
|
GO:0032501
|
multicellular organismal process
|
|
1.32
|
2.87e-07
|
GO:0051239
|
regulation of multicellular organismal process
|
|
2.32
|
3.94e-07
|
GO:0021953
|
central nervous system neuron differentiation
|
|
1.46
|
4.10e-07
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
|
1.23
|
5.84e-07
|
GO:0048523
|
negative regulation of cellular process
|
|
1.52
|
6.34e-07
|
GO:0045944
|
positive regulation of transcription from RNA polymerase II promoter
|
|
1.22
|
6.76e-07
|
GO:0048519
|
negative regulation of biological process
|
|
1.76
|
8.06e-07
|
GO:0002009
|
morphogenesis of an epithelium
|
|
1.53
|
9.74e-07
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
|
1.84
|
9.91e-07
|
GO:0035239
|
tube morphogenesis
|
|
1.43
|
1.48e-06
|
GO:0010628
|
positive regulation of gene expression
|
|
1.91
|
2.40e-06
|
GO:0045165
|
cell fate commitment
|
|
1.78
|
2.46e-06
|
GO:0045664
|
regulation of neuron differentiation
|
|
1.59
|
4.08e-06
|
GO:0007389
|
pattern specification process
|
|
1.42
|
4.60e-06
|
GO:0051254
|
positive regulation of RNA metabolic process
|
|
1.51
|
6.82e-06
|
GO:0023057
|
negative regulation of signaling
|
|
1.36
|
7.16e-06
|
GO:0009888
|
tissue development
|
|
1.24
|
7.53e-06
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
|
1.51
|
8.44e-06
|
GO:0010648
|
negative regulation of cell communication
|
|
2.19
|
1.06e-05
|
GO:0060485
|
mesenchyme development
|
|
1.16
|
1.23e-05
|
GO:0007165
|
signal transduction
|
|
1.48
|
1.24e-05
|
GO:0072358
|
cardiovascular system development
|
|
1.48
|
1.24e-05
|
GO:0072359
|
circulatory system development
|
|
1.52
|
1.60e-05
|
GO:0009968
|
negative regulation of signal transduction
|
|
1.92
|
1.75e-05
|
GO:0030111
|
regulation of Wnt receptor signaling pathway
|
|
1.36
|
2.61e-05
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
|
1.85
|
2.86e-05
|
GO:0060562
|
epithelial tube morphogenesis
|
|
1.46
|
4.80e-05
|
GO:0019226
|
transmission of nerve impulse
|
|
1.46
|
4.80e-05
|
GO:0035637
|
multicellular organismal signaling
|
|
1.31
|
5.43e-05
|
GO:0007154
|
cell communication
|
|
1.19
|
6.57e-05
|
GO:0048522
|
positive regulation of cellular process
|
|
1.99
|
7.20e-05
|
GO:0010720
|
positive regulation of cell development
|
|
1.39
|
8.50e-05
|
GO:0007167
|
enzyme linked receptor protein signaling pathway
|
|
1.33
|
9.56e-05
|
GO:0040011
|
locomotion
|
|
1.47
|
1.53e-04
|
GO:0007268
|
synaptic transmission
|
|
1.26
|
1.84e-04
|
GO:0035556
|
intracellular signal transduction
|
|
1.35
|
2.09e-04
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.18
|
2.13e-04
|
GO:0071842
|
cellular component organization at cellular level
|
|
1.17
|
2.27e-04
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.77
|
2.46e-04
|
GO:0048562
|
embryonic organ morphogenesis
|
|
1.19
|
2.48e-04
|
GO:0051252
|
regulation of RNA metabolic process
|
|
1.76
|
2.70e-04
|
GO:0006813
|
potassium ion transport
|
|
1.85
|
2.70e-04
|
GO:0048705
|
skeletal system morphogenesis
|
|
1.14
|
2.97e-04
|
GO:0031323
|
regulation of cellular metabolic process
|
|
2.04
|
3.17e-04
|
GO:0060828
|
regulation of canonical Wnt receptor signaling pathway
|
|
1.62
|
3.18e-04
|
GO:0048568
|
embryonic organ development
|
|
1.47
|
4.57e-04
|
GO:0000122
|
negative regulation of transcription from RNA polymerase II promoter
|
|
1.49
|
4.84e-04
|
GO:0010551
|
regulation of gene-specific transcription from RNA polymerase II promoter
|
|
1.17
|
4.91e-04
|
GO:0048518
|
positive regulation of biological process
|
|
1.48
|
5.91e-04
|
GO:0043009
|
chordate embryonic development
|
|
1.40
|
6.18e-04
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
|
1.16
|
6.46e-04
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
|
1.48
|
7.18e-04
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
|
1.19
|
7.44e-04
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
|
1.13
|
8.38e-04
|
GO:0051716
|
cellular response to stimulus
|
|
1.41
|
8.83e-04
|
GO:0048585
|
negative regulation of response to stimulus
|
|
1.33
|
9.12e-04
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
|
1.40
|
9.81e-04
|
GO:0007169
|
transmembrane receptor protein tyrosine kinase signaling pathway
|
|
2.06
|
1.06e-03
|
GO:0016331
|
morphogenesis of embryonic epithelium
|
|
2.02
|
1.06e-03
|
GO:0048706
|
embryonic skeletal system development
|
|
1.39
|
1.14e-03
|
GO:0051253
|
negative regulation of RNA metabolic process
|
|
1.50
|
1.15e-03
|
GO:0045597
|
positive regulation of cell differentiation
|
|
1.75
|
1.19e-03
|
GO:0001655
|
urogenital system development
|
|
1.69
|
1.19e-03
|
GO:0030900
|
forebrain development
|
|
1.20
|
1.25e-03
|
GO:0048583
|
regulation of response to stimulus
|
|
1.14
|
1.34e-03
|
GO:0080090
|
regulation of primary metabolic process
|
|
2.07
|
1.71e-03
|
GO:0048762
|
mesenchymal cell differentiation
|
|
2.37
|
1.85e-03
|
GO:0001704
|
formation of primary germ layer
|
|
1.84
|
1.99e-03
|
GO:0043583
|
ear development
|
|
1.30
|
2.07e-03
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
|
1.14
|
2.57e-03
|
GO:0016043
|
cellular component organization
|
|
1.50
|
2.71e-03
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
|
1.38
|
3.21e-03
|
GO:0006935
|
chemotaxis
|
|
1.38
|
3.21e-03
|
GO:0042330
|
taxis
|
|
1.45
|
3.44e-03
|
GO:0060429
|
epithelium development
|
|
1.76
|
3.74e-03
|
GO:0001503
|
ossification
|
|
1.16
|
3.76e-03
|
GO:0010468
|
regulation of gene expression
|
|
1.28
|
4.02e-03
|
GO:0006811
|
ion transport
|
|
1.62
|
4.42e-03
|
GO:0048732
|
gland development
|
|
1.29
|
4.57e-03
|
GO:0009891
|
positive regulation of biosynthetic process
|
|
1.48
|
4.93e-03
|
GO:0051093
|
negative regulation of developmental process
|
|
1.35
|
5.04e-03
|
GO:0010629
|
negative regulation of gene expression
|
|
1.44
|
5.12e-03
|
GO:0051056
|
regulation of small GTPase mediated signal transduction
|
|
1.12
|
5.50e-03
|
GO:0019222
|
regulation of metabolic process
|
|
2.06
|
5.80e-03
|
GO:0014031
|
mesenchymal cell development
|
|
1.40
|
5.94e-03
|
GO:0051094
|
positive regulation of developmental process
|
|
1.15
|
6.03e-03
|
GO:0031326
|
regulation of cellular biosynthetic process
|
|
1.50
|
6.37e-03
|
GO:0001944
|
vasculature development
|
|
1.88
|
6.84e-03
|
GO:0051216
|
cartilage development
|
|
1.96
|
6.86e-03
|
GO:0050769
|
positive regulation of neurogenesis
|
|
1.15
|
7.15e-03
|
GO:0009889
|
regulation of biosynthetic process
|
|
1.61
|
7.49e-03
|
GO:0032582
|
negative regulation of gene-specific transcription
|
|
1.52
|
7.87e-03
|
GO:0045596
|
negative regulation of cell differentiation
|
|
1.40
|
9.01e-03
|
GO:0032583
|
regulation of gene-specific transcription
|
|
1.15
|
9.04e-03
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
|
2.07
|
9.40e-03
|
GO:0043279
|
response to alkaloid
|
|
1.54
|
9.40e-03
|
GO:0003002
|
regionalization
|
|
1.30
|
9.82e-03
|
GO:0007267
|
cell-cell signaling
|
|
1.59
|
1.02e-02
|
GO:0007548
|
sex differentiation
|
|
1.78
|
1.08e-02
|
GO:0060541
|
respiratory system development
|
|
1.74
|
1.09e-02
|
GO:0001763
|
morphogenesis of a branching structure
|
|
1.26
|
1.14e-02
|
GO:0042127
|
regulation of cell proliferation
|
|
1.79
|
1.26e-02
|
GO:0072001
|
renal system development
|
|
2.22
|
1.27e-02
|
GO:0001649
|
osteoblast differentiation
|
|
2.57
|
1.38e-02
|
GO:0061035
|
regulation of cartilage development
|
|
1.49
|
1.47e-02
|
GO:0007423
|
sensory organ development
|
|
1.27
|
1.48e-02
|
GO:0006351
|
transcription, DNA-dependent
|
|
1.82
|
1.67e-02
|
GO:0030323
|
respiratory tube development
|
|
2.05
|
1.97e-02
|
GO:0072175
|
epithelial tube formation
|
|
1.91
|
2.06e-02
|
GO:0030178
|
negative regulation of Wnt receptor signaling pathway
|
|
1.79
|
2.07e-02
|
GO:0001822
|
kidney development
|
|
1.25
|
3.05e-02
|
GO:0009605
|
response to external stimulus
|
|
2.12
|
3.10e-02
|
GO:0090090
|
negative regulation of canonical Wnt receptor signaling pathway
|
|
1.32
|
3.18e-02
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.71
|
3.34e-02
|
GO:0010975
|
regulation of neuron projection development
|
|
1.82
|
3.55e-02
|
GO:0021537
|
telencephalon development
|
|
2.52
|
3.56e-02
|
GO:0021954
|
central nervous system neuron development
|
|
1.77
|
3.75e-02
|
GO:0035107
|
appendage morphogenesis
|
|
1.77
|
3.75e-02
|
GO:0035108
|
limb morphogenesis
|
|
1.80
|
3.96e-02
|
GO:0030324
|
lung development
|
|
2.02
|
4.07e-02
|
GO:0001838
|
embryonic epithelial tube formation
|
|
1.61
|
4.27e-02
|
GO:0010552
|
positive regulation of gene-specific transcription from RNA polymerase II promoter
|
|
1.50
|
4.36e-02
|
GO:0007507
|
heart development
|
|
1.86
|
4.59e-02
|
GO:0021915
|
neural tube development
|
|
1.32
|
4.79e-02
|
GO:0006812
|
cation transport
|
|
1.29
|
4.86e-02
|
GO:0009890
|
negative regulation of biosynthetic process
|
|
1.29
|
4.91e-02
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|