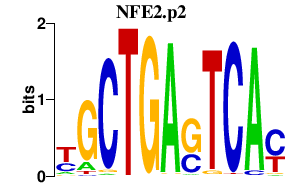
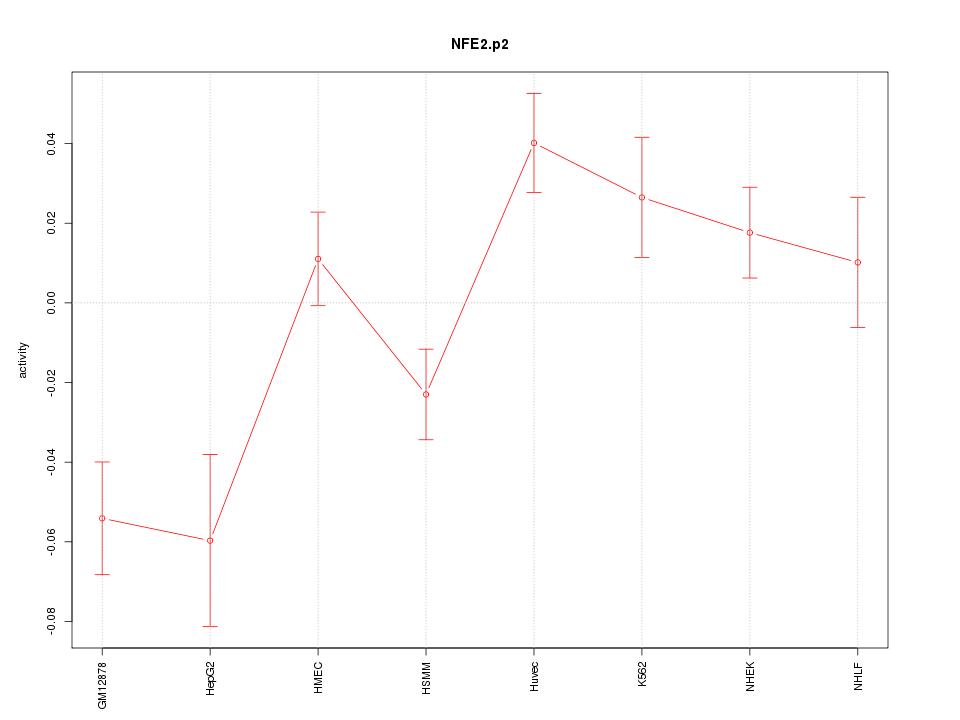
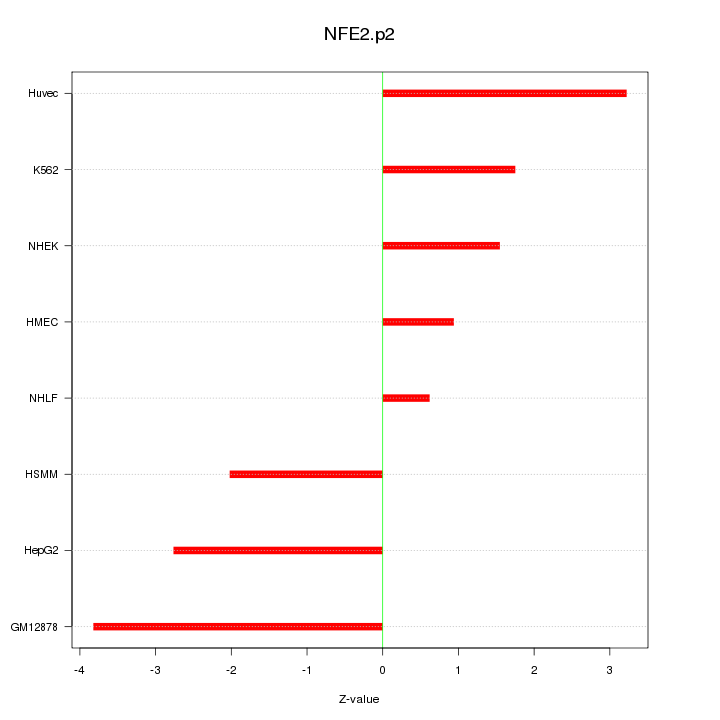
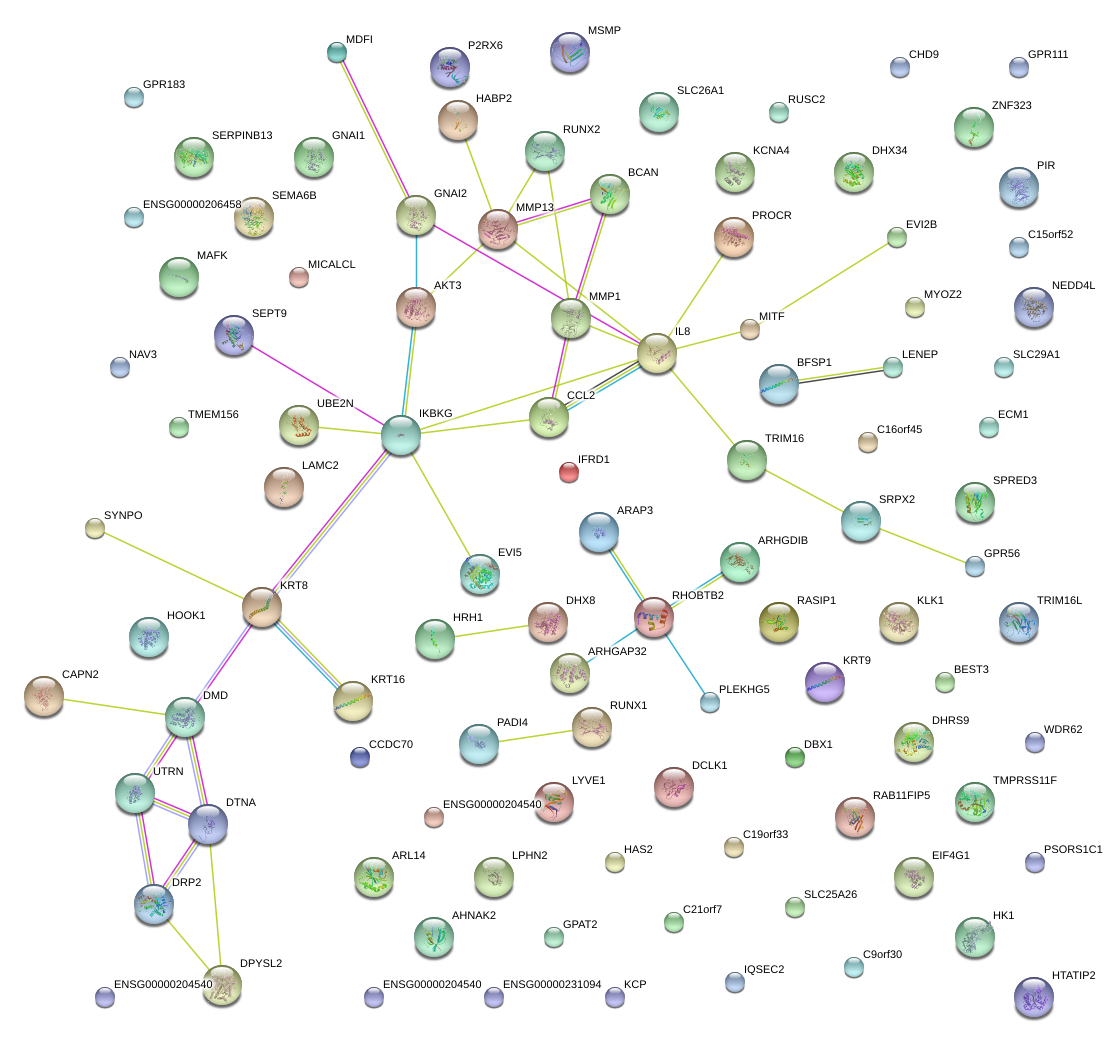
|
chr1_+_221955876
|
14.113
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr12_-_68369322
|
8.414
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr21_+_29439768
|
4.895
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr5_+_150000394
|
4.147
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr5_-_141041982
|
4.094
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr1_+_148747208
|
3.678
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr1_+_148747110
|
3.389
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr9_+_676640
|
3.388
|
|
|
|
|
chr1_+_153232685
|
3.367
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr5_-_141041933
|
3.365
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr1_-_93030538
|
3.352
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr11_-_102174102
|
3.279
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr11_-_102174032
|
3.198
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr17_+_18566060
|
3.183
|
NM_001037330
|
TRIM16L
|
tripartite motif containing 16-like
|
|
chr21_-_35343065
|
2.848
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_+_51334117
|
2.804
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr2_+_169634330
|
2.772
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr6_-_28429950
|
2.759
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr19_+_43572679
|
2.703
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr2_-_73170290
|
2.646
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_+_11153778
|
2.633
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr13_-_35327798
|
2.632
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr9_+_102231616
|
2.625
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr7_+_1543644
|
2.622
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_+_29606408
|
2.592
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chrX_+_153423652
|
2.549
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr9_-_35744271
|
2.436
|
NM_001044264
|
MSMP
|
microseminoprotein, prostate associated
|
|
chr19_-_53935648
|
2.432
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chr12_-_51629874
|
2.412
|
|
KRT8
|
keratin 8
|
|
chr12_+_76749199
|
2.286
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr19_+_43486640
|
2.281
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr18_+_54039779
|
2.281
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr4_+_74825138
|
2.278
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr6_+_41712907
|
2.275
|
|
MDFI
|
MyoD family inhibitor
|
|
chr18_+_54039730
|
2.246
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_15005761
|
2.199
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr19_-_4510713
|
2.198
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr11_+_12265022
|
2.183
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr20_-_17487480
|
2.182
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr16_+_56219802
|
2.177
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr16_+_15503623
|
2.174
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr1_-_242073094
|
2.154
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chrX_+_99786044
|
2.134
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr14_-_104507862
|
2.102
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr3_+_69998064
|
2.086
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr21_-_35343331
|
2.085
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_+_100361588
|
2.026
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr17_-_26665213
|
2.013
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr18_+_54039818
|
2.009
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_+_111850367
|
2.009
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr6_+_44302973
|
1.979
|
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr11_-_128399218
|
1.977
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr3_+_66094324
|
1.923
|
NM_173471
|
SLC25A26
|
solute carrier family 25, member 26
|
|
chr15_-_38417564
|
1.899
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr10_+_70745615
|
1.898
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr17_-_26665194
|
1.888
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_26665144
|
1.845
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr16_+_51690564
|
1.843
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr4_-_68678181
|
1.839
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr1_+_81544432
|
1.821
|
|
LPHN2
|
latrophilin 2
|
|
chr11_-_10546654
|
1.816
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr11_-_102331643
|
1.815
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr1_+_17507276
|
1.793
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr22_+_19699441
|
1.773
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr17_-_37022465
|
1.753
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr18_+_30544193
|
1.743
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr8_+_22900863
|
1.735
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr21_-_35343428
|
1.730
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_31190505
|
1.715
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr4_-_38710376
|
1.715
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr1_-_6474346
|
1.708
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr8_-_122722674
|
1.707
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr3_+_50259329
|
1.703
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chrX_+_99785818
|
1.701
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_+_181421984
|
1.701
|
|
LAMC2
|
laminin, gamma 2
|
|
chr4_-_977163
|
1.670
|
NM_022042
NM_134425
NM_213613
|
SLC26A1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr3_+_161877640
|
1.667
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr10_+_115302767
|
1.660
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr13_-_98757634
|
1.659
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chrX_-_33139349
|
1.641
|
NM_004006
|
DMD
|
dystrophin
|
|
chr3_+_185521139
|
1.622
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr9_+_35528628
|
1.610
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr5_-_90645974
|
1.609
|
|
LOC100505994
|
hypothetical LOC100505994
|
|
chrX_-_53327473
|
1.601
|
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr4_+_120276386
|
1.592
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr11_+_20341806
|
1.559
|
NM_006410
NM_001098521
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr18_+_59405513
|
1.550
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chrX_-_15421279
|
1.542
|
NM_001018109
NM_003662
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr11_+_20341900
|
1.484
|
NM_001098522
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr20_+_33223434
|
1.472
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr7_-_128330615
|
1.464
|
|
KCP
|
kielin/chordin-like protein
|
|
chr21_-_35343448
|
1.462
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_47732181
|
1.452
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr11_+_20341835
|
1.448
|
NM_001098520
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_20342262
|
1.434
|
NM_001098523
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr17_-_36928604
|
1.429
|
NM_002275
|
KRT15
|
keratin 15
|
|
chrX_-_15593074
|
1.419
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr16_+_2023279
|
1.414
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr9_+_35528890
|
1.403
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr14_-_73323629
|
1.399
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr15_+_38849450
|
1.370
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr1_+_12698704
|
1.359
|
NM_001103169
NM_001103170
|
AADACL3
|
arylacetamide deacetylase-like 3
|
|
chr18_+_59593591
|
1.341
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr10_+_47867667
|
1.325
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr5_-_95184121
|
1.320
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr19_-_50977485
|
1.314
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr6_+_32229278
|
1.312
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chrX_-_24575375
|
1.311
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr21_-_35343456
|
1.310
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_101720753
|
1.308
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr7_+_22733290
|
1.305
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr3_+_57069508
|
1.304
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr1_-_110108048
|
1.297
|
NM_024526
NM_133181
NM_139053
|
EPS8L3
|
EPS8-like 3
|
|
chr10_-_31328426
|
1.296
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr17_-_26665220
|
1.286
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_+_181422016
|
1.261
|
|
LAMC2
|
laminin, gamma 2
|
|
chr3_+_185520744
|
1.255
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr8_-_145097031
|
1.249
|
NM_201380
|
PLEC
|
plectin
|
|
chrX_-_24575146
|
1.245
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr6_+_74462226
|
1.241
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr1_-_223683141
|
1.238
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr2_+_113591940
|
1.232
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr9_+_130231027
|
1.198
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr16_+_88515917
|
1.188
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr5_+_35892747
|
1.160
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr3_+_142569988
|
1.154
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr8_-_13018123
|
1.123
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr16_+_65868913
|
1.121
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr4_+_88076418
|
1.116
|
|
|
|
|
chr1_-_159325709
|
1.114
|
|
PVRL4
|
poliovirus receptor-related 4
|
|
chr8_+_24827173
|
1.102
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr4_-_101658094
|
1.100
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr17_+_29621352
|
1.083
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr1_-_151788341
|
1.078
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr12_-_15706340
|
1.077
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr11_-_123685871
|
1.076
|
NM_001002917
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1
|
|
chr1_-_151614681
|
1.057
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr19_-_50977592
|
1.055
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr12_-_101115677
|
1.053
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr2_-_219881937
|
1.042
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr10_+_30763056
|
1.029
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr7_-_150352484
|
1.025
|
NM_173681
|
ATG9B
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chr8_+_24827181
|
1.021
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr18_+_50646616
|
1.016
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr8_-_11748330
|
1.014
|
|
|
|
|
chr19_-_50977586
|
1.013
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr11_-_123261549
|
1.009
|
NM_001013743
|
TMEM225
|
transmembrane protein 225
|
|
chr8_-_82557956
|
1.006
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr5_-_95184221
|
1.006
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr16_+_14187965
|
1.005
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr12_-_55316350
|
1.004
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr19_+_45564549
|
1.004
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr14_-_23802184
|
0.996
|
NM_000359
|
TGM1
|
transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase)
|
|
chr7_-_92300573
|
0.994
|
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr1_-_156923111
|
0.993
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr12_-_25692754
|
0.986
|
NM_001145727
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr11_-_71458739
|
0.983
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr15_+_65245546
|
0.975
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr17_-_43254117
|
0.974
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr19_-_60573552
|
0.974
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr7_-_150561233
|
0.963
|
|
|
|
|
chr12_-_54522881
|
0.959
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr22_-_43986896
|
0.957
|
|
KIAA0930
|
KIAA0930
|
|
chr1_+_181421796
|
0.940
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr7_+_130445394
|
0.938
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr22_+_18388656
|
0.935
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chrX_+_9391314
|
0.926
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr16_+_80626331
|
0.926
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr4_-_159312973
|
0.924
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr11_+_62136788
|
0.923
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr1_+_207668787
|
0.918
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr9_+_4975082
|
0.905
|
|
JAK2
|
Janus kinase 2
|
|
chr4_+_109033868
|
0.903
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_+_49002285
|
0.895
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr18_-_45630154
|
0.892
|
|
MYO5B
|
myosin VB
|
|
chr11_-_62136778
|
0.888
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr11_-_101906640
|
0.872
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr9_+_71009746
|
0.869
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr17_-_43254132
|
0.859
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr1_-_117554999
|
0.851
|
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr15_+_96304911
|
0.832
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr11_-_123353607
|
0.829
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr17_+_25292748
|
0.829
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr9_-_116920232
|
0.828
|
NM_002160
|
TNC
|
tenascin C
|
|
chr1_-_45760115
|
0.824
|
NM_002574
NM_181696
NM_181697
|
PRDX1
|
peroxiredoxin 1
|
|
chr3_-_167037946
|
0.821
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr12_-_51153788
|
0.821
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr5_-_75954807
|
0.816
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr1_-_172258129
|
0.813
|
|
|
|
|
chr18_+_53863865
|
0.811
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_-_85512937
|
0.811
|
NM_013438
NM_053067
|
UBQLN1
|
ubiquilin 1
|
|
chr22_-_43987010
|
0.810
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr12_+_67489063
|
0.809
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr21_-_35343500
|
0.809
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_29563657
|
0.802
|
NM_052967
|
MAS1L
|
MAS1 oncogene-like
|
|
chr1_-_152201814
|
0.799
|
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr17_+_77261477
|
0.789
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr10_-_46601687
|
0.788
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr19_-_55914837
|
0.786
|
|
|
|
|
chr4_-_174687104
|
0.785
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|